Abstract
Transcription factor (TF) binding site (TFBS) models are crucial for computational reconstruction of transcription regulatory networks. In existing repositories, a TF often has several models (also called binding profiles or motifs), obtained from different experimental data. Having a single TFBS model for a TF is more pragmatic for practical applications. We show that integration of TFBS data from various types of experiments into a single model typically results in the improved model quality probably due to partial correction of source specific technique bias.
We present the Homo sapiens comprehensive model collection (HOCOMOCO, http://autosome.ru/HOCOMOCO/, http://cbrc.kaust.edu.sa/hocomoco/) containing carefully hand-curated TFBS models constructed by integration of binding sequences obtained by both low- and high-throughput methods. To construct position weight matrices to represent these TFBS models, we used ChIPMunk software in four computational modes, including newly developed periodic positional prior mode associated with DNA helix pitch. We selected only one TFBS model per TF, unless there was a clear experimental evidence for two rather distinct TFBS models. We assigned a quality rating to each model. HOCOMOCO contains 426 systematically curated TFBS models for 401 human TFs, where 172 models are based on more than one data source.
INTRODUCTION
Regulatory proteins called transcription factors (TFs) bind to DNA sites with specific nucleotide sequences. Many models have been suggested to represent a set of TF binding sites (TFBSs) to which a TF binds (1). The most commonly used model is a position weight matrix (PWM) (2) constructed from a gapless multiple local alignment of TFBSs. The TFBS models are widely used to study transcription regulation in silico. The applications for TFBS models include, for example searching for binding sites and their specific arrangements (3) in putative regulatory sequences, prediction of cis-regulatory modules (4) and detection of possible regulatory role for mutations and genetic variations (5).
Protein binding DNA segments are identified by diverse experimental approaches (6,7), each of which may have a preference for DNA binding segments with specific properties. Typically, these approaches require an additional post-processing involving a TFBS model construction to identify TFBSs more precisely. Different TFs draw different level of attention from experimentalists. Widely used TFBS model collections such as JASPAR (8) and TRANSFAC (9) contain models constructed from hundreds of TFBSs obtained by high-throughput techniques, as well as models constructed from a small number of TFBSs obtained by pre-genomic low-throughput methods. Moreover, for a particular TF, TRANSFAC may contain several models usually derived from data coming from separate experiments, which raises a question which model is the correct one, particularly as they often produce differing predictions.
Models constructed from data obtained with various experimental techniques have different shortcomings. A PWM constructed from a handful of TFBSs obtained by a low-throughput method has less reliable frequencies for non-consensus nucleotides. On the other hand, popular high-throughput methods based on chromatin immunoprecipitation (ChIP) often yield rather long TF bound DNA regions that may contain additional TFBSs for TFs other than the TF under study. When performed in vivo, ChIP also does not allow one to distinguish between direct and indirect binding (10). Therefore, integration of data from both conventional and genome-wide methods may result in a model that would be more reliable than any of the models obtained from single experimental data source.
In most cases, a single ChIP-Seq experiment contains enough information to produce a TFBS binding model. Yet, binding sites found in a particular in vivo experiment provide a condition-specific subset of all putative binding sites. Moreover, it is a common practice to increase specificity by performing motif discovery at a small subset of top binding regions from ChIP-seq data, with weaker bound loci excluded from the analysis, which may further increase the condition-specific bias. A recently forming trend in developing of motif discovery algorithms is to construct complex models (11) assuming a TF can recognize subtypes of binding sites possibly functional in different conditions. Yet, we believe that a generalized TFBS model is still practical for the study of transcription regulation under previously unexplored conditions. Moreover, such models are important as a baseline for comparison of complex models including those accounting for motif subtypes. This is especially important because the increase in model complexity obviously requires more model parameters, which brings about risk of overfitting and complicates estimation of the parameters. Simply speaking, a complex model may entirely miss a binding-site subtype underrepresented in the condition-specific experimental data. Still, such a subtype might be utilized with a simpler model. Another problem of high-throughput data analysis rises from the fact that it normally provides a rather wide binding region, which as a consequence requires considering natural genomic arrangements of binding sites. TFBSs are often located in a complex context of homotypic TFBS clusters (12) or composite elements (9). This makes it particularly difficult to correctly estimate the optimal length of the TFBS model solely based on the high-throughput data compared with the cases when, for example SELEX, in vitro data are used.
The objective of this study is to create a unified and highly non-redundant database of TFBS models, so that each TF is associated with the minimal number of models, while all models are made to have a reliable TFBS recognition quality. In our opinion, such database should be useful in studies of transcription regulation, transcription regulation networks and systems biology applications, where it is of paramount importance to have correct association of a TF to genes it controls. To this end we are integrating binding regions obtained by different experimental methods for a particular TF, which are deposited in different data sources. Contrary to the practice used in compilation of TRANSFAC, we do not integrate experimental data obtained for different TFs that belong to the same TF family.
To create a TFBS model, we used ChIPMunk (13) software for realignment of all binding sequences available for each particular TF. The effectiveness of ChIPMunk has already been confirmed in several independent studies (11,14,15). To improve the quality of the alignment, ChIPMunk allows incorporating a priori information (referred to as ‘a prior’ in what follows). Please note that our approach is not probabilistic and this is not ‘a prior’ in the probabilistic sense. Prior information can include an existing binding site positional preference data such as ChIP-Seq base coverage, that is ChIP-Seq ‘peak shape’ (ChIPMunk terminology: ‘sequence positional prior’) or a model positional prior (ChIPMunk terminology: ‘motif shape prior’) associated with DNA helix pitch. It is notable, that recently a resource providing TFBS models for multiple ChIP-Seq datasets (http://compbio.mit.edu/encode-motifs/) appeared as a spin-off from the ENCODE project (16). Still the analysis was based on traditional motif discovery tools not taking into account specific features of ChIP-Seq data.
Finally, we maintain transparent TF-to-model assignments by having IDs of all resulting models associated with UniProt IDs (17). We emphasize that Homo sapiens comprehensive model collection (HOCOMOCO) is a comprehensive and carefully hand-curated collection of TFBS models with reduced redundancy of model associations to individual TFs. HOCOMOCO contains twice as many TFBS models for human TFs as JASPAR (according to the JASPAR’s public release in 2010), and in average, our models perform better than those from both JASPAR and TRANSFAC in terms of TFBS recognition accuracy (see the Results and Supplementary Material). All our TFBS models, even those constructed from binding sites available from a single data source such as TRANSFAC, are different from existing PWMs (see Supplementary Section 4). We believe that HOCOMOCO complements the existing TFBS model collections such as TRANSFAC and JASPAR and introduces a new promising strategy of TFBS model generation. For TFBS predictions in DNA sequences, we provide predefined PWM score thresholds corresponding to a probability of finding a TFBS among all possible words of a given length. This allows one to have statistically comparable TFBS predictions for various TFBS models in a selected DNA region, which is very convenient for practical purposes. We plan to maintain HOCOMOCO by incorporating additional data as it becomes available and providing an updated set of models with the corresponding annotation. In doing so, our strategy of integrating various experimental data sources allows us to easily incorporate newly available experimental data to update the models or to generate new ones.
MATERIALS AND METHODS
Workflow overview
We started by collecting all available TF binding regions and linking them to the corresponding UniProt IDs. In this way, we have identified 474 TFs with more than 4 available binding regions.
We have shown previously (18) that careful integration of data obtained by different experimental techniques helps to construct more robust TFBS models. We also have shown that the shape of the ChIP-Seq peaks can be effectively used to guide motif discovery (13). In HOCOMOCO, we amended these approaches by taking into account possible model positional priors associated with DNA helix pitch and, when available, the binding site positional preferences.
Sequences of DNA regions binding the same TF and obtained from different independent sources were submitted to ChIPMunk software. For each TF, ChIPMunk was run four times in different computational modes. The four resulting TFBS models were then manually curated to select (what we considered) the best model (or models) for each TF. This was guided by considering known binding preferences listed in UniProt, known TFBS models and models reported for TFs from the same protein family. The overall workflow is presented in Supplementary Section 1. The human curation procedure and criteria are described in specific section later in the text.
DNA protein binding data sources
The initial data on TF binding DNA regions was collected from the following sources: human ENCODE Yale/HudsonAlpha ChIP-Seq (16) presented in the UCSC Genome Browser (19), multiplexed parallel SELEX (20), TRANSFAC 2011.2 SITE table (data for vertebrates) (9) and JASPAR CORE vertebrate (8). In addition, text-mining results based on similar procedure as used in (21,22) and a few manually curated datasets for TFs of particular interest (see Supplementary Section 1 for details) were also utilized as sources.
The procedure of constructing a TFBS model from several data sources implies assigning weights to the sequences from each particular data source before integration. Each type of TF binding data underwent specific pre-processing to select reasonable subset of bound sequences and to weight sequences based on their reliability (e.g. for ChIP-Seq data, the sequence weight was assigned as the peak height). Also sequence positional priors were generated for several datasets, for example using ChIP-Seq base coverage data for Yale ChIP-Seq or according to the graded binding site positions, as given in site annotations of TRANSFAC and JASPAR when available. More details are given in the Supplementary Section 1. To facilitate automatic processing for each TF, all TRANSFAC binding sites available for the particular TF were merged into one dataset. More details on data pre-processing and weighting are given in Supplementary Section 1. Basic statistics for each data source are given in Table 1. Detailed information is given in Supplementary Section 3.
Table 1.
Basic statistics for sequences in each data source
| Data source | Total No. of sequences | Median sequence length (bp) | Average sequence length (bp) |
|---|---|---|---|
| TRANSFAC | 23 199 | 24 | 26 |
| JASPAR | 24 692 | 204 | 207 |
| Yale ChIP-Seq | 96 381 | 454 | 656 |
| HudsonAlpha ChIP-Seq | 65 081 | 559 | 561 |
| Parallel SELEX | 19 535 | 16 | 16 |
| All other datasets | 2655 | 1000 | 687 |
Assigning weights to data sources
For TFs with several independent sources of DNA binding data, the following empirical procedure was used to integrate data for TFBS model generation similar to that used for motif discovery in (18). Let there be k data sources with Nk binding sites in each, making N binding sequences in total. Each dataset receives the weight of Wk, where Wk = C log (Nk + 1), and C is selected in a way that Sum(Wk) = N. For each kth dataset, the weight Wk is then proportionally distributed between sequences according to their sequence weights so the sum of sequence weights in the kth dataset equals to Wk. The correct weighting of initial data is very important. Even for sequence analysis of huge sets from ChIP-Seq data, different motif discovery tools produce similar but not the same TFBS models from the same data (see, e.g. comparison from completeMOTIFs pipeline, http://cmotifs.tchlab.org/help.html). For huge sequence sets and well-defined TFBS models, the effects of weighting schema variation can be comparable with those produced by usage of different motif discovery methods. Our weighting schema is selected heuristically in a way that small datasets (like SELEX or footprints) play a supporting role in identification of similar TFBS models in large datasets (ChIP-Seq) but do not substantially influence the final motifs. If a too large weight is assigned to a small source dataset, then the resulting TFBS model depends heavily on the removal of a single TFBS from this small dataset that appears undesirable. Still, if a small sequence set is the only one available, then these heuristics can produce artifact TFBS models. We hope that human curation helps to overcome this issue. An overview of the weighting scheme is presented in Supplementary Section 2a.
TFBS model identification modes
To construct TFBS models, ChIPMunk was run four times: two times (f1) and (f2) with uniform model positional prior and two times (si) and (do) with informative model positional prior (see the next section).
Using a weighted set of N sequences as the input data, ChIPMunk searches for an optimal model in a given range of gapless local multiple alignment lengths. The min-to-max (f1) and max-to-min (f2) model length estimation modes were used with the minimum length of 7 bp and the default maximum length of 25 bp. The maximum tested alignment length was reduced in a number of cases when input sequences were not long enough (see details in Supplementary Section 2e). Supplementary Section 2e also presents a description of ChIPMunk parameters used during the TFBS model construction procedure.
Informative model positional priors
It was observed that in TFBS alignments, the positional information content is often modulated in accordance with a DNA helix pitch (23). The suggested explanation is that better aligned nucleotides contact the protein bound by the major groove (24). Although, there are proteins binding the minor groove or forming other types of structural complexes with DNA, binding by the major groove is the most common (25). This property was also taken into account in HeliCis (26), where a Gibbs sampler was used assuming a periodic prior for spacing between boxes of double box motif.
For ChIPMunk, we used the single (si) and double box (do) model positional priors. For a single box, the positional weights are to be distributed as cos2 (πn/T), where T = 10.5 is the DNA helix pitch, n is the coordinate within the alignment and the center of the alignment is at n = 0. During the internal cycle of PWM optimization (see Supplementary Section 2c–d), the PWM column scores are multiplied by prior values so the columns closer to the center of the alignment (n = 0) receive no score penalty while the columns around (n = 5, 6, −5, −6) contribute much less to the score of the PWM under optimization. The single box model prior was used along with the min-to-max length estimation mode (si). We also used the double box model prior with a shape prior equal to sin2(πn/T), which was used to search for possibly longer double box models in the max-to-min length estimation mode (do). The presented priors do not cover all possible arrangements of DNA-protein binding because there are TFs, which bind with variable spacers between boxes like P53 or TFs specifically interacting with the DNA minor groove such as TBP. Thus, the hand curation step is introduced to select the most reasonable model for each particular case.
Selection of an appropriate model
In many cases, four ChIPMunk modes yielded convincingly similar resulting models for self-consistent datasets and well-defined TF binding models (such as CTCF or REST).
Our ultimate objective was to create a TFBS model collection with low redundancy. So of four TFBS models for each TF, it was necessary to select the most appropriate one. The selection procedure was based on the following ideas. If we got a stable model (identical optimal gapless multiple local alignments) using two or more ChIPMunk modes, then we considered this model as obtained in the simplest possible mode (with the flat shape prior or with the single box prior whatever was available). When all modes yielded different models, we selected the mode having the highest alignment weight (i.e. the total weight of all sequences included in the resulting alignment) unless the model built from fewer sequences was closer to known models from the same TF family or the known consensus listed in UniProt. When a sequence set was small (no more than a few dozens of binding sites), we usually selected the shortest available model containing presumably only core positions (positions with the high information content) as the flanking nucleotides are most likely less reliable.
In 25 cases, 2 models were selected for 1 TF (Supplementary Table S1a). This occurred if the TF was shown to bind DNA either as a monomer and a homodimer or as homodimer and heterodimer preferentially with TFs from one particular group having a similar binding model (according to information listed in UniProt).
Criteria for the model quality assignment
The resulting models were rated according to their quality; the ratings were assigned by human curation according to the following criteria:
Relevant distribution of position-specific information content over alignment columns, which means a model LOGO representation displaying well-formed core positions with a high-information content surrounded by flanking positions with lower information content; the information content at flanking positions decreasing with the distance from the model core.
‘Stability’, which means that in more than one of the ChIPMunk modes, we obtained models with a similar length, consensus and comparable number of aligned binding sites, along with a similar shape of model LOGO representation.
‘Similarity’ of the model to the binding sequence consensus for this TF given in the UniProt or other databases, which means similarity of the shape of the model LOGO and TFBS lengths to those of other TFs from the same TF family.
‘A total number of binding sites’ was also considered as a quality measure, because a large set of binding regions (mostly but not limited to ChIP-Seq and parallel SELEX) implies that there are many observations of each nucleotide in any position of the alignment, particularly many observations of non-consensus nucleotides in core positions. In low-information content positions, where there is no strong consensus, all variants have many observations, so the observed nucleotide frequencies are less sensitive to fluctuations. If a set had more than a hundred sequences, we considered it large. However, sometimes, it was impossible to produce a model complying with requirements 1–3 even from a very large set, so we never considered the volume of the data as a principal criterion.
Model quality assignment
One of six quality rates, from A to F, was assigned to each model. Model quality rates from A-to-D were assigned to proteins known to be TFs, including those listed in (27) with addition of a number of proteins having relevant models and sufficient evidence to be TFs. Quality A was assigned to models complying to all four criteria listed in the section above. Quality B was assigned to models built from large sequence sets that failed no more than one of the three remaining criteria. Quality C was assigned to models built from small sequence sets but (with a number of specifically marked exceptions) complying with the three remaining criteria. Quality D models missed part of the known consensus sequence or had no clearly significant core positions in the TFBS model. Quality E (error) was assigned to models for proteins not convincingly shown to be TFs or to models exhibiting a non-specific LOGO shape or a wrong consensus sequence (comparing to known UniProt consensus). Quality F (failure) was assigned to TFs for which no reliable model was identified. Details on curation and quality assignment are provided in Supplementary Table S1. Full information on TFBS models of different quality is available on the HOCOMOCO website.
PWM thresholds
For each model, we produced the list of pre-computed PWM thresholds showing corresponding P-value, that is a fraction of all 4l words (where l is the word length, i.e. the PWM width) scoring above the threshold. Such thresholds allow one to normalize TFBS predictions for different TFs, so that the same PWM hit rate is expected in a random sequence (similar to false positive); this is very convenient for practical purposes. The P-values were calculated by MACRO-APE software (http://autosome.ru/macroape/) using the strategy described in (28). For each predefined P-value level and the corresponding threshold, we also show the percentage of all aligned words (finally included in the optimal alignment) scoring no less than the threshold and the percentage of initial binding segments (used during motif discovery) having PWM hits scoring no less than the threshold.
Assessing TFBS recognition quality
In (18), we have provided a comprehensive assessment of data integration strategy based on different types of conventional low-throughput and ChIP-chip data. ChIP-Seq data gives many advantages over ChIP-chip, so it is especially important to check how our pipeline was performing on such datasets for human TFs. To this end, we used a strategy similar to that in (29). We selected 36 TFs with ChIP-Seq data available and their available TFBS models in TRANSFAC and JASPAR databases. For these TFs, we produced independent positive control sequence sets made of up to 1000 ChIP-Seq peaks not involved in estimation of HOCOMOCO model parameters. For a true positive (TP), we adopted a case when a peak from the positive control set contained at least one PWM hit scoring no less than the threshold.
For each PWM, we reordered sequences from the positive control set according to their maximal PWM hit scores. The resulting decreasing set of PWM scores was considered as a set of PWM threshold values, where each threshold corresponded to a particular TP rate value.
For each PWM threshold, we computed the Ps as the probability to finding at least one PWM hit with a score no less than the threshold in a random double-strand DNA segment of a fixed length L. For L, we selected the median length of the sequences in the positive control set for the TF in consideration.
Ps was calculated from the PWM P-value P of obtaining a given score for a random word at the particular position of a random double-strand DNA sequence (calculated as in (28), see PWM Thresholds above) assuming the hits (including overlapping hits) being independent and their number complying compound Poisson distribution:
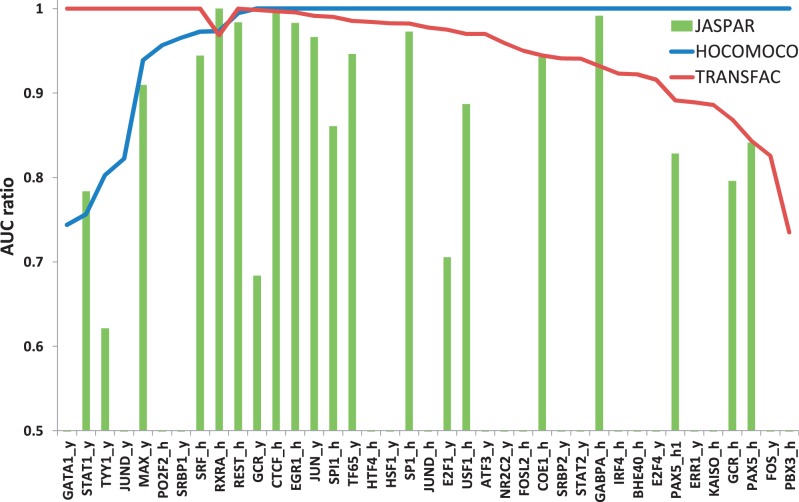
To assess the recognition quality of the models, we plotted a set of receiver operating characteristic (ROC) curves showing the TP rate versus the Ps (being estimation for a false-positive rate) for a set of PWM thresholds based on the positive control set. ROC curves are given in Supplementary Section 5. Higher curves correspond to models with better TFBS recognition quality. To quantitatively estimate model quality, we have calculated area-under-curve (AUC) values. Thus, for each TF, we compare HOCOMOCO, JASPAR (when available) and all appropriate TRANSFAC TFBS models. The AUC values are presented in Supplementary Table S3. To compare performances of TRANSFAC, JASPAR and HOCOMOCO for each TF, we computed the AUC value for all its models and then divided these values by the largest AUC value obtained for these models producing a ratio value. So the best model for each TF always received the ratio value of 1. The result is displayed in Figure 1, with TFs ordered according to their increasing HOCOMOCO AUC ratios.
Figure 1.
Comparison of AUC ratios for TFBS models of JASPAR (green bars), TRANSFAC (red curve) and HOCOMOCO (blue curve) TFBS models. Value of 1 corresponds to the best model with the highest AUC value. Points on X-axis correspond to control sets for different TFs. Y-axis shows AUC ratios. If several TFBS models were present in a collection, the best result is shown. Details are given in the text.
RESULTS AND DISCUSSION
The HOCOMOCO websites provide initial binding sequence data (but no binding sites from TRANSFAC), alignments and position count/weight matrices and precomputed matrix threshold levels. HOCOMOCO-AD collection includes 426 binding models of quality A–D for 401 TFs. Twenty-five TFs have TFBS sets described by two models. For the sake of completeness, we also provide the E- (55 models) and full- (1896 models for 474 TFs) collections. The average TFBS length of A–D models is 12 bp. Table 2 shows an overview of the models of different quality ratings.
Table 2.
Overview of the HOCOMOCO TFBS models of different quality ratings
| Quality | TFs | Models | Sequences per TF (median) | Data sources per TF (median) |
|---|---|---|---|---|
| A | 49 | 52 | 2037 | 2 |
| B | 82 | 87 | 159.5 | 2 |
| C | 128 | 139 | 38.0 | 1 |
| D | 142 | 148 | 16.5 | 1 |
| E | 55 | 55 | 11.0 | 1 |
| F | 18 | — | 2047.0 | 1 |
Manual annotation of 18 proteins with F-quality models reveals 5 TFs with probable sequence-specific binding, 3 putative TFs and 10 non-TF proteins (see Supplementary Table S2 for details). In most cases, the F quality assignment was indeed characteristic for proteins not binding specifically to DNA and thus having no clearly visible DNA recognition preferences. This agrees with the expectation that no reasonable model should be obtained when sequences in the dataset contain no common sequence signal.
At the P-value of 0.0005, models of HOCOMOCO-AD collection in average recognize more than 80% of words used to construct PWMs. On average, more than 75% of the initial sequences have PWM hits above the 0.0005 P-value thresholds.
To check TFBS recognition quality, we have applied the strategy discussed in Materials and Methods to compare TRANSFAC and JASPAR TFBS models with HOCOMOCO based on the independent positive control set. The AUC values are presented in Supplementary Table S3. The graphic comparison of AUC ratios is shown in Figure 1. Suffixes ‘_y’ and ‘_h’ correspond to Yale and HudsonAlpha ChIP-Seq datasets, respectively. For PAX5, there were two HudsonAlpha datasets (‘_h’ and ‘_h1’) based on different antibodies. HOCOMOCO models show better quality in 29 of 39 cases (more than 70%). JASPAR performs the best in a single case (RXRA). It is notable that TRANSFAC versus HOCOMOCO comparison shows different results on Yale versus HudsonAlpha for JUND and GCR TFs. In almost all cases, TRANSFAC contained several models that could correspond to different TFBS subtypes, in such cases only the best model is shown in comparison, the resulting multiple testing effect in this case favored performance of TRANSFAC compared with that of JASPAR or HOCOMOCO.
In more than half of the cases, the manually selected TFBS models were obtained in two or more ChIPMunk modes. Thus, theoretically, it is possible to construct an automated heuristic procedure that would perform similar to human curation at least for half of TFs. Still, when different runs resulted in dissimilar, or similar but not the same models, the manual curation was necessary to select an appropriate model. Moreover, in some cases, the manually selected model corresponded to shorter TFBS sequences than the stable model, especially when the sequence set was small.
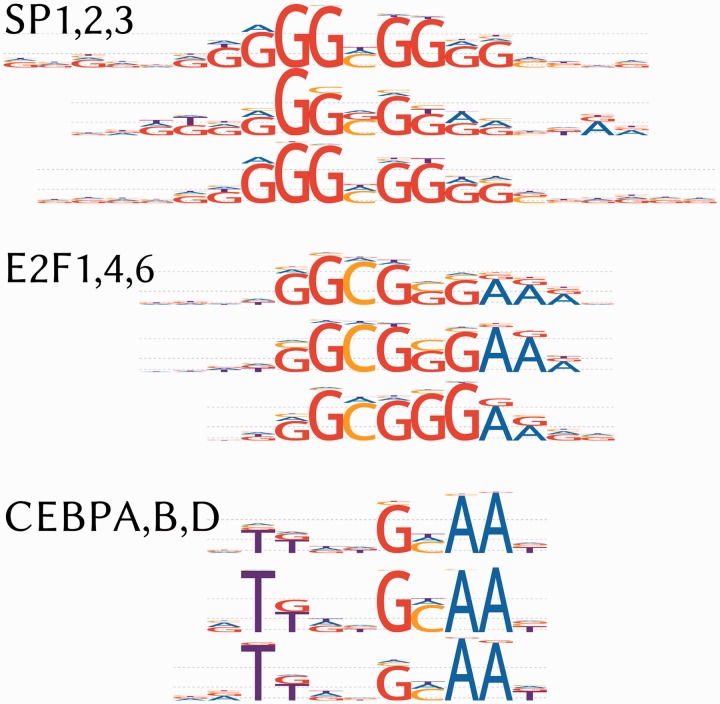
In most of the cases, it was possible to select models with similar consensuses and LOGO shapes for TFs from the same family (e.g. see CEBP, E2F or Sp families; LOGOs are given in Figure 2). We constructed a pairwise similarity matrix for HOCOMOCO models of highest quality (A–C) using Jaccard similarities computed with the help of MACRO-APE software (http://autosome.ru/macroape/). The matrix was then supplied to the UPGMA (30) clustering procedure resulting in a hierarchical tree. An interactive representation of this tree can be used as another way to browse HOCOMOCO collection. It is notable that several known families of TFs form tight clusters providing an indirect validation of our manual curation procedure.
Figure 2.
TFBS model LOGOs for highly similar models within TF families. LOGOs for selected members of CEBP, E2F and SP families are given. The Discrete Information Content is used for nucleotide scaling as in (29). Note that in our LOGO representation, the dominant nucleotides are placed at the bottom enabling easy observing the sequence of the best scoring binding site.
Because many HOCOMOCO models were based on the sequence sets taken from TRANSFAC only, we additionally demonstrated that all our models are novel in a sense that they recognize TFBSs different from those detected by TRANSFAC PWMs. To this end, we computed the minimal Jaccard distance (1 – similarity) between HOCOMOCO-AD and TRANSFAC models. The results (see Supplementary Section 4) demonstrated that the HOCOMOCO and TRANSFAC models indeed displayed some similarity (which one could expect because they corresponded to the same TFs), but the typical Jaccard distance between the closest HOCOMOCO and TRANSFAC models was usually greater than ∼0.7 at the PWM P-value of 0.0005. This means that the words recognized in parallel by the PWM from HOCOMOCO and the PWM from TRANSFAC at the corresponding threshold levels amounted to less than 30% of words recognized by any of two PWMs. There were no PWM pairs with zero or close to zero distances. The minimal distance between HOCOMOCO and TRANSFAC PWM was greater than 0.2 corresponding to more than 20% of words being recognized by only one of two PWMs.
The use of priors based on DNA structural properties in 25 cases allowed us to produce 2 distinct high-quality models, each of which was supported by the ability of a given TF to bind DNA as monomer or homodimer/heterodimer. In 31 of 50 cases, we succeeded in obtaining good quality models from the Yale ChIP-Seq sets profiting from the base coverage (peak shape) available for this dataset. It is noteworthy that those models had A/B quality in 25/6 cases, respectively, no models of C or D quality, 7 models of E quality and 12 models with F quality that corresponded mostly to non-TF proteins (see Supplementary Table S2). For the ChIP-Seq data with no information on base coverage (HudsonAlpha ChIP-Seq), we had 14/6/1/2/2/4 of A/B/C/D/E/F quality models, respectively. Most of the models with A–C quality had additional data sources.
As one can see from Table 2, the suggested quality assignment procedure resulted in reasonable ratings having higher quality models based on a greater number of data sources. The only exception is the set of F quality models mostly based on ChIP-Seq data. The possible reason for this quality assignment is that some of ChIP-Seq data are related to the non-TF proteins, so such proteins do not bind DNA in a sequence-specific manner or bind DNA only in a complex with other proteins. Other possible explanations for low-quality ChIP-Seq–based models also include indirect or non-specific DNA binding of a TF, unknown experimental bias in a particular ChIP-Seq experiment, and inadequacy of the gapless local multiple alignment or its PWM representation as a TFBS model.
Overall, we have provided a resource that contains hand-curated TFBS models for human TFs based on variety of binding data available and using a number of criteria aimed to increase the quality of resulting models. Our comparison analysis of models from two other major TFBS model resources (TRANSFAC and JASPAR) convincingly demonstrates that our strategy and the resulting models are performing with improved performance quality in most cases as expected. Thus, HOCOMOCO represents a useful complement of TRANSFAC and JASPAR databases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Sections 1–5 and Supplementary References [31,32].
FUNDING
Dynasty Foundation Fellowship [to I.V.K.]; Russian Foundation for Basic Research [12-04-32082-mol_a to I.V.K.]; Presidium of the Russian Academy of Sciences Program in Cellular and Molecular Biology; Presidium of the Russian Academy of Sciences Fundamental Research Subprogram ‘Gene pools dynamics and conservation’; Russian Ministry of Science and Education State Contract [07.514.11.4005]; Russian Ministry of Science and Education State Contract [07.514.11.4006]; Russian Ministry of Science and Education grant [11.G34.31.0008]. Funding for open access charge: Presidium of the Russian Academy of Sciences program in Cellular and Molecular Biology.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Trafica, LLC and personally Ivan Lysov for providing computational resources. We also thank Evolutionary Genomics Laboratory, Faculty of Bioengineering and Bioinformatics, M.V. Lomonosov Moscow State University and personally Prof. A.S. Kondrashov for providing additional computational facilities. We specially thank Arieh Sherris for his many important suggestions on the manuscript.
REFERENCES
- 1.Bailey TL. Discovering sequence motifs. Methods Mol. Biol. 2008;452:231–251. doi: 10.1007/978-1-60327-159-2_12. [DOI] [PubMed] [Google Scholar]
- 2.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 3.Kulakovskiy IV, Belostotsky AA, Kasianov AS, Esipova NG, Medvedeva YA, Eliseeva IA, Makeev VJ. A deeper look into transcription regulatory code by preferred pair distance templates for transcription factor binding sites. Bioinformatics. 2011;27:2621–2624. doi: 10.1093/bioinformatics/btr453. [DOI] [PubMed] [Google Scholar]
- 4.Nikulova AA, Favorov AV, Sutormin RA, Makeev VJ, Mironov AA. CORECLUST: identification of the conserved CRM grammar together with prediction of gene regulation. Nucleic Acids Res. 2012;40:e93. doi: 10.1093/nar/gks235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macintyre G, Bailey J, Haviv I, Kowalczyk A. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010;26:i524–i530. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elnitski L, Jin VX, Farnham PJ, Jones SJ. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- 7.Geertz M, Maerkl SJ. Experimental strategies for studying transcription factor-DNA binding specificities. Briefings Funct. Gen. 2010;9:362–373. doi: 10.1093/bfgp/elq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi Y, Kim H, Gupta R, Davuluri RV. Tree-based position weight matrix approach to model transcription factor binding site profiles. PLoS One. 2011;6:e24210. doi: 10.1371/journal.pone.0024210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulakovskiy IV, Boeva VA, Favorov AV, Makeev VJ. Deep and wide digging for binding motifs in ChIP-Seq data. Bioinformatics. 2010;26:2622–2623. doi: 10.1093/bioinformatics/btq488. [DOI] [PubMed] [Google Scholar]
- 14.Kuttippurathu L, Hsing M, Liu Y, Schmidt B, Maskell DL, Lee K, He A, Pu WT, Kong SW. CompleteMOTIFs: DNA motif discovery platform for transcription factor binding experiments. Bioinformatics. 2011;27:715–717. doi: 10.1093/bioinformatics/btq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X, Kulkarni A, Zhang Z, Xuan Z, Serfling R, Zhang MQ. A highly efficient and effective motif discovery method for ChIP-seq/ChIP-chip data using positional information. Nucleic Acids Res. 2012;40:e50. doi: 10.1093/nar/gkr1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulakovskiy IV, Makeev VJ. Discovery of DNA motifs recognized by transcription factors through integration of different experimental sources. Biophysics. 2010;54:667–674. [Google Scholar]
- 19.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr1055. (Database issue), D918–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpää MJ, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajic VB, Veronika M, Veladandi PS, Meka A, Heng MW, Rajaraman K, Pan H, Swarup S. Dragon plant biology explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists. Plant Physiol. 2005;138:1914–1925. doi: 10.1104/pp.105.060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan H, Zuo L, Choudhary V, Zhang Z, Leow SH, Chong FT, Huang Y, Ong VW, Mohanty B, Tan SL, et al. Dragon TF association miner: a system for exploring transcription factor associations through text-mining. Nucleic Acids Res. 2004;32:W230–W234. doi: 10.1093/nar/gkh484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papp PP, Chattoraj DK, Schneider TD. Information analysis of sequences that bind the replication initiator RepA. J. Mol. Biol. 1993;233:219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- 24.Schneider TD. Consensus sequence Zen. Appl. Bioinformatics. 2002;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- 25.Pabo C, Sauer R. Protein-DNA recognition. Annu. Rev. Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 26.Larsson E, Lindahl P, Mostad P. HeliCis: a DNA motif discovery tool for colocalized motif pairs with periodic spacing. BMC Bioinformatics. 2007;8:418. doi: 10.1186/1471-2105-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer U, Schmeier S, Bajic VB. TcoF-DB: dragon database for human transcription co-factors and transcription factor interacting proteins. Nucleic Acids Res. 2011;39:D106–D10. doi: 10.1093/nar/gkq945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Touzet H, Varré JS. Efficient and accurate P-value computation for position weight matrices. Algorithms Mol Biol. 2007;2:15. doi: 10.1186/1748-7188-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulakovskiy IV, Favorov AV, Makeev VJ. Motif discovery and motif finding from genome-mapped DNase footprint data. Bioinformatics. 2009;25:2318–2325. doi: 10.1093/bioinformatics/btp434. [DOI] [PubMed] [Google Scholar]
- 30.Sokal R, Michener C. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958;38:1409–1438. [Google Scholar]
- 31.Abramowitz M, Stegun IA. Handbook of Mathematical Functions. New York, NY: Dover Publications; 1972. [Google Scholar]
- 32.Lifanov AP, Makeev VJ, Nazina AG, Papatsenko DA. Homotypic regulatory clusters in Drosophila. Genome Res. 2003;13:579–588. doi: 10.1101/gr.668403. [DOI] [PMC free article] [PubMed] [Google Scholar]