Abstract
The availability of comprehensive information about enzymes plays an important role in answering questions relevant to interdisciplinary fields such as biochemistry, enzymology, biofuels, bioengineering and drug discovery. At the EMBL European Bioinformatics Institute, we have developed an enzyme portal (http://www.ebi.ac.uk/enzymeportal) to provide this wealth of information on enzymes from multiple in-house resources addressing particular data classes: protein sequence and structure, reactions, pathways and small molecules. The fact that these data reside in separate databases makes information discovery cumbersome. The main goal of the portal is to simplify this process for end users.
INTRODUCTION
The number of registered databases in the NAR Database Issue during the last year has increased 7% to 1380 (1) with coverage of a great variety of fields and scopes. Within this landscape, users usually have to navigate from one resource to another in order to gather the information they need. That is where portals designed for a particular community can play an important role, simplifying the search process and presenting views of data from different databases.
Many portals already exist which integrate data from different resources. Biomart Central Portal (2) uses the generic Biomart federation technology to present combined data from virtually any biology database. BioPortal (3) gives access to biomedical ontologies and tools to work with them. BioProject and BioSample (4) include metadata annotations in their registers, which allow the aggregated search of experimental data submitted to different NCBI, EBI and DDBJ databases.
Other portals are focused on more specific scientific fields. The Technology Portal of PSI-SBKB (5) integrates technology summaries and tools for structural biology, along with videos and social networks. InterStoreDB (6) provides plant phenotypic and genomic data from different sequence, crop and alignment databases. The IKMC web portal is a friendly interface to search many different BioMarts for data about targeted and trapped mouse knockout availability and structure (7).
Enzymes—biomolecules that catalyse specific chemical reactions—are of central importance in many fields of science, and their study is the foundation of the entire field of biochemistry. Enzymes have key regulatory and metabolic roles and are of large commercial and healthcare importance. The engineering of new functions, and the development of specific inhibitors for enzymes, is becoming more important in the post-genomic era. The EBI hosts a number of resources providing data on enzymes. One of them is IntEnz, the Integrated Enzyme relational database that provides the NC-IUBMB nomenclature and classification and facilitates downloads in different formats. Other EBI databases that provide enzyme data, such as UniProtKB and PDBe, are cross-referenced from IntEnz. However, these hyperlinks do not provide a good overview of the information users require and make it very difficult for users to navigate. In addition, there are other resources—Reactome, ChEBI, ChEMBL, Rhea—containing detailed data about the biochemistry of enzymes which could also be integrated for the benefit of users. It was with these aims in mind that the enzyme portal was launched.
The enzyme portal is a free resource that summarizes publicly available information about enzymes, including small-molecule chemistry, biochemical pathways and drug compounds. It provides a concise summary of information from:
UniProt Knowledgebase (8);
Protein Data Bank in Europe (PDBe) (9);
Rhea, a database of enzyme-catalysed reactions (10);
Reactome, a database of biochemical pathways (11);
IntEnz, a resource with enzyme nomenclature information (12);
ChEBI (13) and ChEMBL (14), which contain information about small-molecule chemistry and bioactivity;
MACiE (15) for highly detailed, curated information about reaction mechanisms;
EFO (16), the Experimental Factor Ontology, a system for annotation of experiments from which the enzyme portal retrieves disease-related information, concretely from children of its ‘disease’ entry.
The enzyme portal collates diverse information about enzymes and displays it in an organized overview. It covers many species, including mammals, invertebrates and plants, and provides a simple way to compare orthologues.
MATERIALS AND METHODS
User-centred design was applied to improve usability and meet the expectations of users
The portal was designed by following a user-centred design lifecycle (17), whereby decisions regarding the design of the portal were made based on evidence gathered from users representing our target audience, namely, scientists working in enzyme-related research, such as biomarker discovery, enzymology and drug discovery (see also Supplementary Methods).
The portal was implemented using Java technologies
We used Java technologies—Spring Web MVC, JAXB, JAX-WS—and several Web Service APIs either SOAP—EB-Eye, ChEBI, CiteXplore—or REST—UniProt, Reactome, Rhea, ChEMBL, BioMart, DAS—to build the web application. XML schemas describe the underlying model.
For quick cross-referencing and building filters for search results, an Oracle database was populated with cross-references to/from UniProtKB identifiers to other databases, including those not directly cross-referenced currently in the UniProt Knowledgebase such as ChEBI and ChEMBL. UniProtKB accessions are used throughout the enzyme portal as enzyme identifiers.
SEARCHING THE ENZYME PORTAL
Searches in the enzyme portal are based on the powerful EB-eye search engine (18), which indexes many EBI resources, updating for every release and provides a web service API.
From the website homepage, a free text search can use any relevant query terms, such as enzyme names, EC numbers, UniProtKB accessions, gene names and small-molecule names.
Search results are shown in a table (Figure 1) with orthologues grouped and split into separate pages where appropriate. The results list the enzyme name, a description of its function and any synonyms as well as the list of species in which the enzyme is found. If there are any related diseases, these will also be displayed.
Figure 1.
Search results are grouped as orthologues. Note the paging (top right of the table) and the filter facets (left).
Each result shows on its left side a fully coloured thumbnail of the protein structure or a greyed image if none is found for any of the grouped orthologues.
Search filters
On the left-hand side, there is a list of species, compounds and diseases which are related to the search results. Users can filter these simply by clicking the corresponding checkbox so that only enzymes matching the checked items will be shown, hiding the others.
Species
A list of species where enzymes included in the search results are found is shown so that users can narrow the search to any specie(s) of interest (Figure 2).
Figure 2.
Search results for ‘CFTR’ filtered to show only enzymes known in Ma’s night monkey.
Compound
Any small molecules known to interact in some way with the enzymes in the list are shown here and can also be used as filters. This includes cofactors, activators, inhibitors and drugs.
Disease
Search results can also be filtered according to any diseases associated to them. For example, checking a box labelled ‘stroke’ will display only those enzymes which have been related to this disease.
Note that several filters within the same section—species, compounds or diseases—have the effect of union of results (boolean OR), while filters from different sections result in intersection of results (boolean AND). For example, checking Xenopus laevis and Drosophila melanogaster will display only enzymes present in one species or the other; checking Rattus norvegicus and GMP3- will display only enzymes present in rat and known to interact with the nucleotide.
ENZYME DATA
Clicking on an enzyme name (or one of its orthologues) in the list of search results takes to the enzyme page, organized into tabs (on the left-hand side).
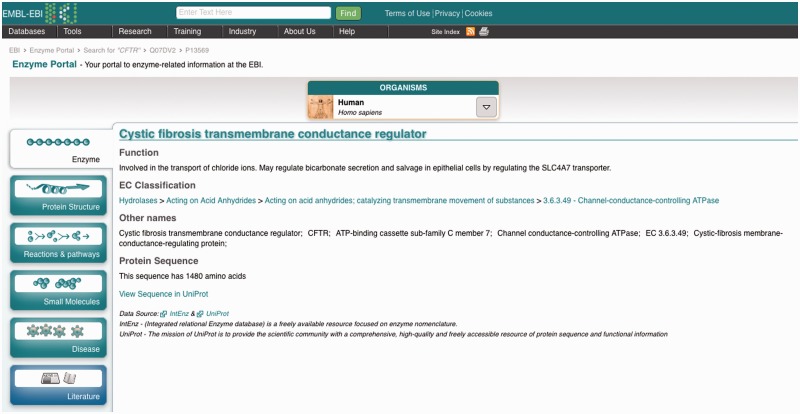
Enzyme summary
The first one (Figure 3) is the enzyme summary tab, which contains the description of the enzyme function, its classification in the EC hierarchy, any synonyms and some information about the protein sequence. An ‘organisms’ drop-down menu at the top can be used to switch between the known orthologues of the enzyme, from any of the available tabs.
Figure 3.
Enzyme summary. Orthologues can be selected using the ‘organisms’ drop-down menu at the top. Notice the breadcrumbs at the top left, which include links to the search results and any other orthologues visited previously.
While navigating search results and orthologues, a history of the users’ navigation is kept in the form of breadcrumbs.
Throughout the portal, all data sources are acknowledged and linked, so that users can access in-depth information (such as the protein sequence from UniProtKB or the enzyme EC classification from IntEnz).
Protein 3D structure
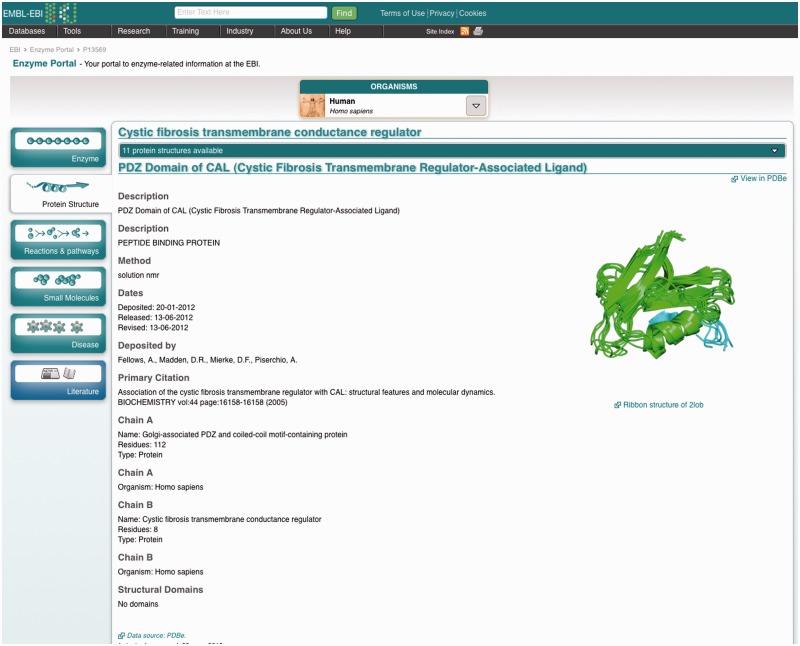
The protein structure tab (Figure 4) shows any experimental 3D models of the enzyme. If there are several of them, any one can be selected from the drop-down menu, which indicates the number of structures available.
Figure 4.
3D structure tab showing basic information about experimental models of the protein structure.
Only basic information on the model is displayed: again, users can navigate to the PDBe web pages that describe the structure.
Reactions and pathways
The reactions and pathways tab (Figure 5) shows the biochemical reaction(s) catalysed by the enzyme—with the chemical structures of the participants linked to ChEBI—as well as any metabolic pathways in which the enzyme may be involved.
Figure 5.
Reactions and pathways tab: chemical structures are hyperlinks to the corresponding entities in ChEBI. Additional information about the reaction available from Rhea, Reactome and MACiE is also linked from here. This tab also includes a list of pathways in with every reaction can be involved, including descriptions and graphics when available.
Only summarized information is shown in this tab. For more information—additional data about the reaction, reaction mechanisms and context within pathways—users are referred to the data source (in this case, Rhea, MACiE and Reactome, respectively) with the provided hyperlinks when available.
Small molecules
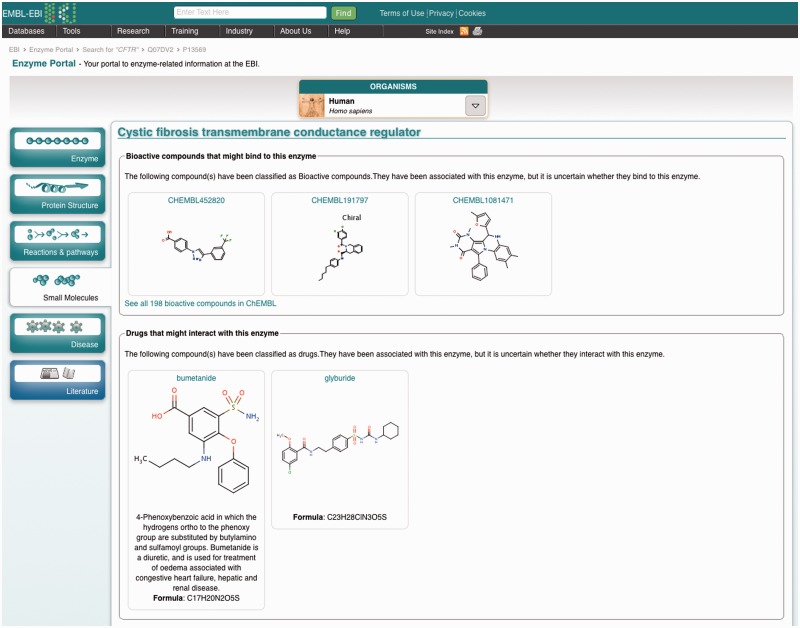
The small molecules tab (Figure 6) includes any available information from UniProtKB about cofactors, activators, inhibitors and drugs, and also any bioactive compounds from the ChEMBL database which have been associated with the enzyme. The chemical structures are links to navigate to ChEBI (with its focus on structure and nomenclature) or ChEMBL (with its focus on bioactivity and function).
Figure 6.
Small molecules that have been associated in some way to the enzyme, including drugs, inhibitors and activators.
Diseases
The disease tab (Supplementary Figure S1) lists any diseases that are related to the selected enzyme—OMIM entries and MeSH terms cross-referenced from UniProtKB, translated into EFO identifiers, with a short description of the disease and how it relates to the enzyme.
Literature
The literature tab (Supplementary Figure S2) lists bibliographic citations relevant to the enzyme. Article titles are linked to the EBI’s CiteXplore bibliography database. If an abstract is available, clicking on the ‘toggle abstract’ link will show it.
Citations can be filtered according to the different aspects of the enzyme, i.e. the other tabs.
DISCUSSION
The enzyme portal integrates many resources, most of them hosted by EBI and also external ones such as BioPortal. Its main goal is to provide information about enzymes in a suitable format, with a usable interface designed for intended users. Instead of reinventing the wheel, it makes use of available and reliable resources to that end.
Although some of these resources already incorporate real semantics, others do not, which makes it difficult to extract meaningful information on enzymes using existing semantic web technologies. This portal fills these gaps using the well-known relationships between the EBI databases.
The EB-eye tool (18) provides gene and protein summaries for search results. The enzyme portal complements this with additional metabolic information: catalytic activity, pathways, regulation by small molecules and related diseases. It also keeps a look and feel consistent with that of the EB-eye summaries. The original data sources are always acknowledged and linked from the enzyme portal pages. This portal offers useful overviews on enzymes, but users are referred to the original databases in order to get in-depth information.
The data provided by the enzyme portal are live in the sense that they are not stored in a data warehouse, but retrieved on demand from the different data sources. This way, maintenance is reduced to a minimum and the most recent data are guaranteed by relying on web services from the provider databases, which is an advantage over the data warehouse approach.
The enzyme portal is a one-stop shop for enzyme-related information in resources developed at the EBI. It accumulates this information and aims to present it to the scientist with a unified user experience. The enzyme portal team does not curate enzyme information and therefore is a secondary information resource or portal. At some point, a user interested in more detail will always leave its pages and refer to the information in the underlying primary database (UniProtKB, PDBe, etc.) directly.
BRENDA (19) is the most comprehensive resource about enzymes worldwide and has invested a great amount into the abstraction and curation of enzymes and their related information. BRENDA contains valuable information that cannot be found in the enzyme portal at the moment, such as substrate, kinetic, specificity, stability, application, disease-related and engineering data. As a primary resource, BRENDA could be a candidate for an information source for the enzyme portal in the future.
Future technical developments
Programmatic access through web services will be provided in the future, making use of the existing XML schemas defining the underlying object model. Other features demanded during the design process were customized downloads—users will be able to save search results and enzyme summaries in the format of their choice—and side-by-side enzyme comparison. Additional means to browse the data through enzyme classification, compound classification and disease annotation has also been requested.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2, Supplementary Methods and Supplementary References [20–22].
FUNDING
The European Molecular Biology Laboratory (core funding). Funding for open access charge: European Molecular Biology Laboratory—European Bioinformatics Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank the invaluable help received from all the participants involved in the user-centred design process (workshops and user testing) for the design of the portal and also from the data providers.
REFERENCES
- 1.Galperin MY, Fernández-Suárez XM. The 2012 nucleic acids research database issue and the online molecular biology database collection. Nucleic Acids Res. 2012;40:D1–D8. doi: 10.1093/nar/gkr1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guberman JM, Ai J, Arnaiz O, Baran J, Blake A, Baldock R, Chelala C, Croft D, Cros A, Cutts RJ, et al. BioMart Central Portal: an open database network for the biological community. Database. 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musen MA, Noy NF, Shah NH, Whetzel PL, Chute CG, Story MA, Smith B, NCBO team The National Center for Biomedical Ontology. J. Am. Med. Inform. Assoc. 2012;19:190–195. doi: 10.1136/amiajnl-2011-000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gifford LK, Carter LG, Gabanyi MJ, Berman HM, Adams PD. The Protein Structure Initiative Structural Biology Knowledgebase Technology Portal: a structural biology web resource. J. Struct. Funct. Genomics. 2012;13:57–62. doi: 10.1007/s10969-012-9133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Love CG, Andongabo AE, Wang J, Carion PW, Rawlings CJ, King GJ. InterStoreDB: a generic integration resource for genetic and genomic data. J. Integr. Plant Biol. 2012;54:345–355. doi: 10.1111/j.1744-7909.2012.01120.x. [DOI] [PubMed] [Google Scholar]
- 7.Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Kadin JA, Bult CJ, Eppig JT, Oakley DJ, Briois S, et al. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39:D849–D855. doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Golovin A, Gore SP, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2012;40:D445–D452. doi: 10.1093/nar/gkr998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcántara R, Axelsen KB, Morgat A, Belda E, Coudert E, Bridge A, Cao H, de Matos P, Ennis M, Turner S, et al. Rhea—a manually curated resource of biochemical reactions. Nucleic Acids Res. 2012;40:D754–D760. doi: 10.1093/nar/gkr1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haw R, Stein L. Using the reactome database. Curr. Protoc. Bioinformatics. 2012 doi: 10.1002/0471250953.bi0807s38. Chapter 8, unit8.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcántara R, Ast V, Axelsen KB, Darsow M, de Matos P, Ennis M, Morgat A, Degtyarenko K. IntEnz. Molecular Biology Database Collection entry number 508. Nucleic Acids Res. 2007 [Google Scholar]
- 13.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, et al. ChEMBL: a large-scale bioactivity database for chemical biology and drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holliday GL, Andreini C, Fischer JD, Rahman SA, Almonacid DE, Williams ST, Pearson WR. MACiE: exploring the diversity of biochemical reactions. Nucleic Acids Res. 2012;40:D783–D789. doi: 10.1093/nar/gkr799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an experimental factor ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavelin K, Cham JA, de Matos P, Brooksbank C, Cameron G, Steinbeck C. Bioinformatics meets user-centred design: a perspective. PLoS Comput. Biol. 2012;8:e1002554. doi: 10.1371/journal.pcbi.1002554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valentin F, Squizzato S, Goujon M, McWilliam H, Paern J, López R. Fast and efficient searching of biological data resources—using EB-eye. Brief. Bioinform. 2010;11:375–384. doi: 10.1093/bib/bbp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:670–676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt J, Adlin T. The Persona Lifecycle: Keeping People in Mind Throughout Product Design. San Francisco, CA: Morgan Kaufmann; 2006. [Google Scholar]
- 21.Gray D, Brown S, Macanufo J. Game Storming: A Playbook for Innovators, Rulebreakers, and Changemakers. Sebastopol, CA: O’Reilly; 2010. [Google Scholar]
- 22.Snyder C. Paper Prototyping: The Fast and Easy Way to Design and Refine User Interfaces. San Francisco, CA: Morgan Kaufmann; 2003. [Google Scholar]