Abstract
UCNEbase (http://ccg.vital-it.ch/UCNEbase) is a free, web-accessible information resource on the evolution and genomic organization of ultra-conserved non-coding elements (UCNEs). It currently covers 4351 such elements in 18 different species. The majority of UCNEs are supposed to be transcriptional regulators of key developmental genes. As most of them occur as clusters near potential target genes, the database is organized along two hierarchical levels: individual UCNEs and ultra-conserved genomic regulatory blocks (UGRBs). UCNEbase introduces a coherent nomenclature for UCNEs reflecting their respective associations with likely target genes. Orthologous and paralogous UCNEs share components of their names and are systematically cross-linked. Detailed synteny maps between the human and other genomes are provided for all UGRBs. UCNEbase is managed by a relational database system and can be accessed by a variety of web-based query pages. As it relies on the UCSC genome browser as visualization platform, a large part of its data content is also available as browser viewable custom track files. UCNEbase is potentially useful to any computational, experimental or evolutionary biologist interested in conserved non-coding DNA elements in vertebrates.
INTRODUCTION
Several comparative studies on whole vertebrate genomes uncovered non-coding sequences that exhibit extremely high conservation. DNA regions of perfect identity shared between human, mouse and rat with length >200 bp have been called ultraconserved elements (UCEs) (1). Similar DNA regions have been referred to by others as conserved non-genic regions (2), conserved non-coding elements (CNE) (3) or highly conserved non-coding elements (HCNE) (4). The exact number of such elements depends on the criteria used for their identification, see (5) for review. The multitude of different names may appear unfortunate and confusing. Nevertheless, we decided to use yet another term, ultraconserved non-coding elements (UCNE), to make clear that our resource is restricted to the most highly conserved class of such elements and excludes protein-coding regions.
Although the strong conservation of these sequences points to an important biological role, a known molecular mechanism that would require such a high degree of conservation is currently unknown. Experimental studies in transgenic animals suggest that most of these sequences act as tissue-specific enhancers during developmental processes (6–8). A striking property of vertebrate UCNEs is that they cluster in genomic regions containing genes coding for transcription factors and developmental regulators (so called trans-dev genes) (4,9). These clusters show conserved synteny between distant genomes and are called ‘genomic regulatory blocks’ (GRBs) (10).
The experimental characterization of UCNEs faces limitations. For instance, it is currently not possible to study molecular interactions of UCNEs within single cells of a developing organism. In the absence of adequate experimental techniques, comparative genomics approaches represent a promising alternative to gain some clues about their function. Understanding how UCNEs have evolved in the past may tell us something about what they do today.
Here, we present UCNEbase, a comprehensive resource on the genomic organization and evolution of vertebrate UCNEs and ultraconserved genomic regulatory blocks (UGRBs). We will first explain the procedures by which the database was built, before we describe its contents and the user interfaces. A comparison of UCNEbase with already existing resources on CNEs will be presented in the ‘Discussion’ section.
DATABASE CONCEPT AND DATA ACQUISITION
UCNEbase provides information on the evolution and genomic organization of 4351 UCNEs in multiple vertebrate species. Around half of these elements are located within intergenic regions (2139) and the rest are located within non-coding parts of genes: introns (1713) and UTRs (499). As most UCNEs occur as arrays near key trans-dev genes, our resource is organized along two hierarchical levels: individual UCNEs and UGRBs.
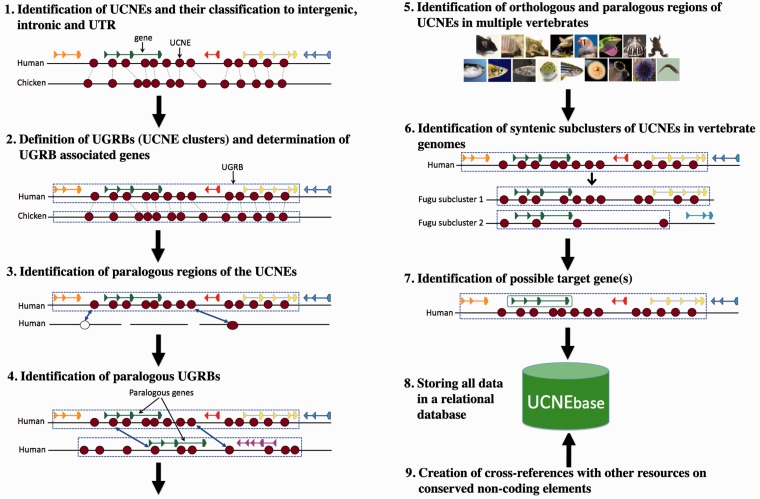
The information provided by UCNEbase is generated by a combination of automatic procedures and manual curation steps. The methodology used for the creation of UCNEbase is schematically shown in Figure 1. A brief description of each step follows below. Technical details are provided in the Supplementary Methods.
Figure 1.
Schematic representation of the methodology used for the creation of UCNEbase.
Definition of UCNEs
We defined UCNEs as non-coding human DNA regions that exhibit ≥95% sequence identity between human and chicken and are >200 bp. The sequence identity threshold corresponds to a base substitution rate of ∼1% per 100 million years. We have previously shown that sequences fulfilling such stringent criteria exist only in vertebrates (11). To compile a list of human UCNEs, we scanned whole-genome alignments between human and chicken downloaded from UCSC (12) with a sliding window technique. Human and chicken were selected as reference species for two main reasons: (i) their evolutionary distance provides high specificity in detecting functional elements (13) and (ii) both genome assemblies are of high quality and thus suitable for identifying large syntenic regions. From the initially extracted set of ultra-conserved sequence elements, we eliminated coding regions and a few human repetitive sequences aligning with the same chicken sequence. The remaining 4351 sequences composed our reference set of UCNEs. Each element of this set was then classified as either ‘intergenic’, ‘intronic’ or ‘UTR associated’ according to the human gene annotation from RefSeq. The length of the UCNEs identified in this way ranged from 200 to 1419 bp with a mean 325 bp and a median 283 bp. The total length is 1.4 Mb.
The criteria used to identify UCNEs are admittedly arbitrary, like any other criteria used before. In particular, there is no objective boundary between UCNEs and HCNEs. However, the primary goal of UCNEbase is not to be a comprehensive resource. It should rather be considered an exploratory tool to study the general features of UCNEs with the aid of a stringently selected collection of prominent examples.
Definition of UGRBs
We defined ‘UGRBs’ (also referred to as ‘UCNE clusters’) as arrays of UCNEs that are syntenically conserved between the human and chicken genomes. Syntenic conservation means that the orthologues of the individual UCNEs of a human UGRB occur in the same order within a restricted area of a chicken chromosome. During the initial scan, we required that neighbouring UCNEs must not be separated by >0.5 Mb in both human and chicken. However, a few exceptions to this rule were made during subsequent manual curation based on visual inspections of the genomic context. Currently, UCNEbase comprises 239 UGRBs encompassing 3868 UCNEs. The number of UCNEs within a UGRB varies considerably from 134 in the ZEB2 cluster to only 2 in the ONECUT2 cluster (with an average of 16 and a median of 8 UCNEs). The genomic size of the identified UGRBs also varies significantly from 4.9 Mb (IRXB cluster) to ∼2 kb (CPEB4 cluster).
For each UGRB, we defined a corresponding set of UGRB-associated genes comprising all genes that fall within or overlap with a genomic region spanned by the block. If the UGRB starts or ends with an intergenic UCNE, the upstream and downstream flanking genes were also included. The set UGRB-associated genes was sometimes expanded during subsequent manual curation steps, for instance by including paralogues of genes from a paralogous UGRB.
Identification of human UCNE paralogues
Human genomic regions that exhibit significant sequence similarity to a UCNE are considered paralogues of that UCNE. In general, CNEs have fewer paralogues compared with protein-coding genes. However, the relatively rare cases of UCNE paralogues are highly informative with regard to the origin of UGRBs. Most paralogous UCNEs originate from ancient whole-genome duplication events that happened at the root of the vertebrate tree (14).
As we do not expect high sequence conservation between UCNE paralogues, we compared each UCNE against the complete human genome (split into overlapping pieces of 10 kb) using the program SSEARCH v34 from the FASTA package (15). This program is an implementation of the sensitive Smith–Waterman local alignment algorithm. The initial scan was performed with a permissive E-value threshold. We subsequently re-evaluated the statistical significance of each match by computing base composition-adjusted E-values using the classical window shuffling test (16). All matches with E-value ≤10−4 were retained as paralogues.
Our systematic search for paralogues confirmed the expectation that most UCNEs are unique. Only 464 UCNEs have at least one human paralogue. Of the 1252 paralogous regions found, only 177 were UCNEs themselves.
Identification of paralogous UGRBs
We also tried to identify paralogous relationships at the level of GRBs. This was mostly done by manual curation. As a minimal condition, we required that two UGRBs share at least one paralogous gene. However, most paralogous blocks also share paralogous UCNEs. In some cases, synteny across paralogous blocks was used to redefine the extension of individual UGRBs. In total, 82 UGRBs were found to have at least one paralogous UGRB forming 39 groups.
Detection of UCNE orthologues in other species
Currently, UCNEbase contains information about UCNE homologues (orthologues and paralogues) in 18 vertebrate genome assemblies that include four mammals: mouse (mm10), armadillo (dasNov1), opossum (monDom5) and platypus (ornAna1); two birds: chicken (galGal3) and zebra finch (taeGut1); two reptiles: lizard (anoCar2) and painted turtle (chrPic1); one amphibian: Xenopus (xenTro3) and five fishes: fugu (fr2), medaka (oryLat2), stickleback (gasAcu1), Tetraodon (tetNig2) and zebrafish (danRer7). We also identified a few UCNEs (mainly located in UTRs) that have orthologues in lamprey (petMar1), Ciona intestinalis (ci2), sea urchin (strPur2) and lancelet (braFlo1). To identify these homologues, we performed Smith–Waterman searches against the complete genomes using the same protocol as for the identification of paralogues within the human genome.
Once the homologous regions were defined, we classified each of these regions as either an orthologue or a paralogue. In doing so, we made the assumption that a homologue of a human UCNE could be an orthologue of the same UCNE or an orthologue of one of its paralogous regions in the human genome. To distinguish between these two cases, we compared each homologue of a human UCNE with all human paralogues of that UCNE (if there were any). If a better alignment score (lower E-value) was obtained with a paralogue, then the UCNE homologue was classified as paralogue. If the alignment scores were very close, we visually inspected the corresponding genomic regions and based our judgement on orthology annotation for nearby genes or synteny with other UCNE homologues.
Identification of syntenic subclusters of UCNEs in vertebrate genomes
For each human UGRB, we identified ‘orthologous syntenic subclusters’ of UCNEs in other vertebrates. An orthologous syntenic subcluster is a set of UCNE orthologues that occurs as a cluster on the same chromosome, scaffold or contig in another vertebrate genome assembly such that any two neighbouring UCNEs are separated by ≤0.5 Mb. For most species, we would expect only one orthologous cluster per UGRB. In reality, we often find one cluster plus a few isolated orthologous UCNEs located on sequence contigs not assigned to chromosomes. The situation could be different in the five fish species that have undergone a lineage-specific whole-genome duplication (see example in Figure 1).
Identification of possible target genes
GRBs are generally assumed to control only one target gene belonging to the so-called trans-dev family. With all the information on orthologous and paralogous regions in other genomes at hand, we tried to identify the most likely target gene for each cluster. To this end, we primarily relied on a genomic context analysis approach. We reasoned that target genes will always be conserved together with UCNEs after whole-genome duplication events. Based on the analysis of the gene content of paralogous UGRBs in human and the fate of UGRB-associated UCNEs in duplicated fish genomes, we were often able to identify a single target gene. In the cases where we were left with several candidates, we gave preference to genes encoding transcription factors. In fact, the overwhelming majority of target genes uniquely defined by genomic context analysis turned out to be transcription factors, most of them containing either zinc fingers or homeodomains, or both.
Perhaps, we could have used transcriptional and epigenetic features as well to single out the most likely target genes from multiple candidates, as suggested by a recent study (17). Technically, however, it is not obvious how this should be done. We are currently exploring several ways how such experimental data could be exploited for target gene identification in the future.
UGRB and UCNE nomenclature
One of the distinctive features of UCNEbase is the establishment of a coherent nomenclature for UCNEs that ensures that orthologous UCNEs have the same names in all species. These names may also serve as unique identifiers for UCNEs, which is a real issue because nowadays non-coding elements are often referred to by genomic coordinates relating to a specific genome assembly. If the corresponding genome assembly is not mentioned, the location of the element will no longer be traceable in a few years from now.
In UCNEbase, we try to define names that carry some information about the function and genomic location of a UCNE, as well as its evolutionary relationship to other UCNEs. UCNE names are typically composed of two parts: a UGRB name and an element name. For example, DACH1_Ava and DACH1_Benjamin are two UCNEs that belong to the DACH1 cluster. UGRBs have the same name as their putative target genes. Elements are identified by common people’s names or names from mythology. Within a UGRB, the alphabetical order of the elements reflects the linear arrangement of the elements along a chromosome. Importantly, paralogous UCNEs share the same element name (e.g. DACH1_Hana is a paralogue of DACH2_Hana). For elements that are not part of a UGRB, the corresponding chromosome name replaces the block name, e.g. chr2_Nemo. The rule that paralogues should have the same name extends to non-clustered UCNEs (e.g. chr10_Sherlock is a paralogue of CPEB2_Sherlock).
A small number of UCNEs are very close to each other and thus could be part of the same functional entity. To specifically mark such cases, UCNEs that are separated by ≤50 bp in both human and chicken are given the same element name, however extended by different serial numbers (e.g. DACH1_Scheherazade_1 and DACH1_Scheherazade_2). Note further that our naming scheme is extensible by design. If we were to add a new UCNE to an existing UGRB in the future, it will be easy to find a name that alphabetically fits between the two neighbouring UCNEs.
CONTENT AND USER INTERFACES
As UCNEbase is organized along two hierarchical levels, there are two types of entries, UCNEs and UGRBs. About 90% of UCNE entries are related to UGRBs. There is only one entry per UCNE or UGRB, containing information for all vertebrate species covered by the resource. UCNEbase is organized in a human-centric fashion. Each entry type has two parts: one providing detailed information relating to the human genome and a second part providing information on homologous elements and clusters of elements in other species. There is a standard html display for each entry with internal links to paralogous conserved regions and external links to other databases and genome browsers. The UCSC genome browser is used as the major visualization platform. A large part of the information contained in UCNEbase is provided as browser-viewable BED files.
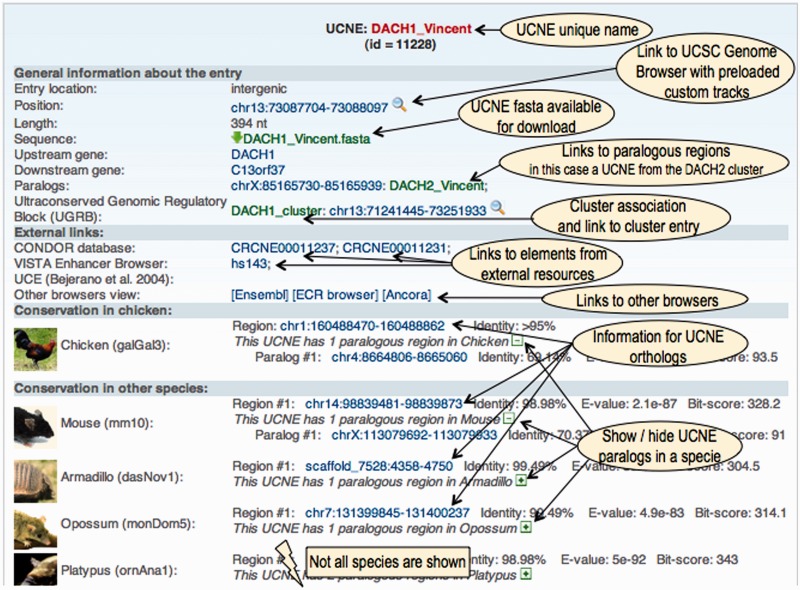
Content of a UCNE entry
The first part of a UCNE entry contains the following data items:
a unique name;
the location relative to the nearest genes (intergenic, intron or UTR);
the genome coordinates in UCSC format;
the length of the UCNE;
the sequence in FASTA format;
the names of overlapping genes (for intronic and UTR-associated UCNEs), or the nearest upstream and downstream genes (for intergenic UCNEs);
a list of human paralogous UCNEs (identified by name and genomic coordinates) and other paralogous regions (identified by genomic coordinates only);
the name of the corresponding UGRB (if any);
cross-references to overlapping entries from the CONDOR database (18), VISTA Enhancer Browser (19) and Bejerano’s UCE collection (1).
The second part contains information about homologous regions in other vertebrates. The regions are defined by genomic coordinates and classified as either orthologues or paralogues. In addition, the sequence identity, E-value and bitscore of the local alignments are stored.
A web display of a UCNE entry is shown in Figure 2. Note that all genomic coordinates are linked to the UCSC genome browser through hyperlinks that automatically pre-load a number of custom tracks from UCNEbase. The web display also provides links to the Ancora (20), ECR (21) and Ensembl (22) genome browser. Some information contained in a UCNEs entry will not automatically be displayed. The DNA sequence is only accessible through a hyperlink. Under the section header ‘Conservation in other species’, only orthologous regions are displayed initially. The paralogous regions can be made visible by clicking on the ‘+’ button.
Figure 2.
Web display of a UCNE entry.
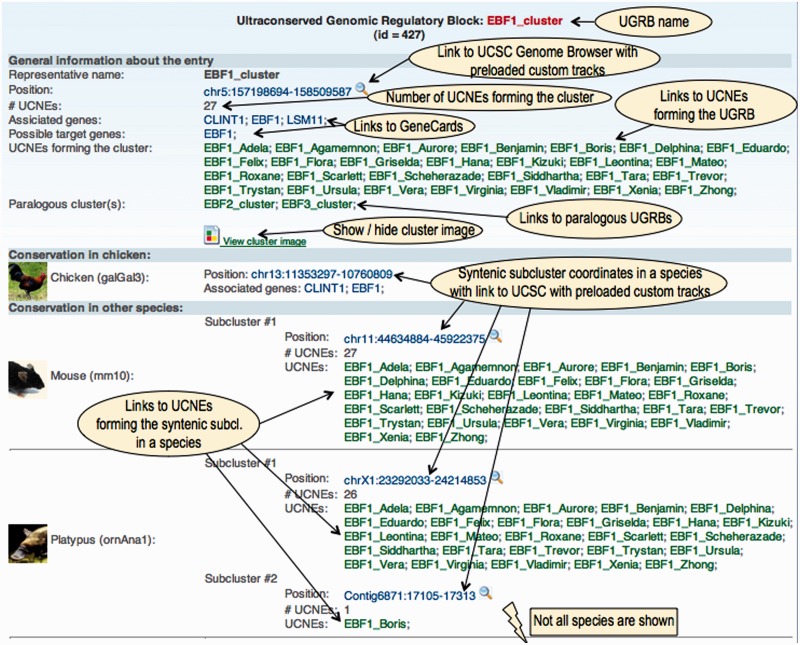
Content of a UGRB entry
The main section of a UGRB entry contains the following data items:
a unique name corresponding to the most likely target gene;
the genome coordinates in UCSC format;
the number of UCNEs forming the block;
a list of all human genes associated with the block;
a list of possible target genes (in most cases only one);
a list of all UCNEs forming the block;
a list of paralogous UGRBs;
The section on sequence conservation contains synteny maps of UGRBs across multiple vertebrate genomes. A complete synteny map for a given species consists of one or several syntenic blocks referred to as ‘subclusters’. The information associated with a subcluster comprises the genomic coordinates, the number of orthologous UCNEs and the names of these UCNEs.
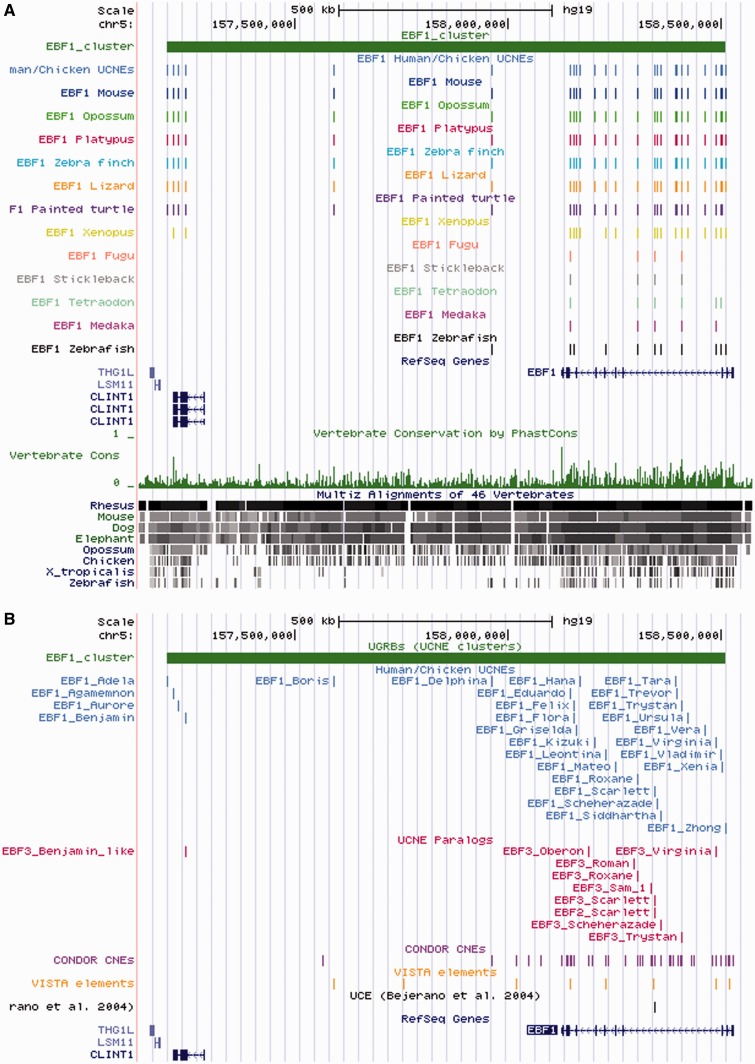
An example of a web display of a UGRB entry is shown in Figure 3. As for UCNE entries, the genomic coordinates are all linked to the UCSC genome browser and will automatically pre-load a number of custom tracks. The web display also includes a locally stored image (a UCSC browser snapshot) providing an overview of orthologous UCNEs in different vertebrate genomes (Figure 4A). It is initially hidden, but can be made visible through an on–off button. Note that the image shows only a part of the information contained in the custom tracks provided by UCNEbase. A mouse click on the image will open a UCSC genome browser window, in which the tracks can be explored in more detail (Figure 4B).
Figure 3.
Web display of a UGRB entry.
Figure 4.
UCSC browser view of the EBF1 cluster with custom tracks from UCNEbase. (A) Summary picture of cross-genome conservation provided by UCNEbase. (B) Detailed view of the ‘human/chicken UCNEs’ and ‘UCNE paralogues’ tracks accompanied by a dense view of the tracks indicating conserved elements from other resources.
Data access and visualization
UCNEbase provides several query mechanisms to find UCNEs and UGRBs based on different search criteria. All entries can be accessed by their chromosomal location in the human genome or by proximity to particular genes through the web links ‘Browse UCNE clusters’ and ‘Browse individual UCNEs’. The ‘Advanced search’ page allows searches by additional criteria, including genomic location in other vertebrate species. Yet another page provides access through external database IDs from the CONDOR database, VISTA Enhancer Browser and Bejerano’s UCEs collection.
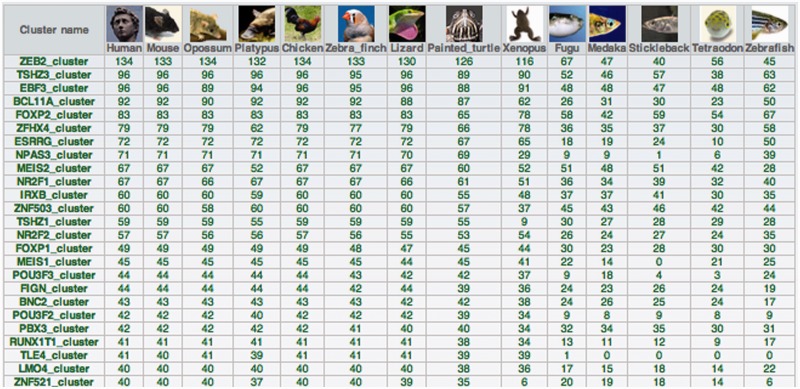
UCNEbase also provides three fully hyperlinked summary tables, one containing a list of paralogous UGRBs, another one containing a list paralogous UCNEs and a third one (entitled ‘species cluster summary’) showing the numbers of conserved UCNEs for each UGRB in all species (Figure 5).
Figure 5.
Species cluster summary. The table shows the number of orthologous UCNEs found in different genomes for the 25 largest UGRBs.
UCNEbase relies on the UCSC genome browser for data visualization. A large part of the information content is available as custom track files. This has the principle advantage that information from UCNEbase can be explored together with a great variety of genome annotations from other sources. The UCSC browser also serves as a navigation platform. All data items from UCNEbase that can be displayed in a browser window are back-linked to the corresponding UCNE and UGRB entries. For instance, in the example shown in Figure 4B, clicking on the blue box labelled ‘EBF1_Oberon’ will take the user back to corresponding UCNEbase entry. From there, one could use a link back to the genome browser to view an orthologous UCNE from another species.
For the human genome, UCNEbase provides custom tracks for UCNEs, UGRBs, UCNE paralogues, CONDOR CNEs, Vista elements and UCEs from Bejerano’s collections. In addition, there is a group of tracks showing the subset of UCNEs conserved in different species. For non-human species, there are tracks for UCNE orthologues, UCNE paralogues and subclusters of UCNEs corresponding to human UGRBs.
DISCUSSION
There are several other resources on CNEs with partially overlapping objectives, in particular: CONDOR, CORG (23), cneViewer (24), Ancora, VISTA Enhancer browser, ECR browser and TFCONES (25). Despite a common theme, the scopes of these resources are quite different which makes a direct comparison difficult. For instance, a significant portion of CONDOR and the VISTA Enhancer browser consists of experimental annotation of non-coding elements based on in vivo reporter gene assays in zebrafish and mouse. Such information is not within the scope of UCNEbase. Other resources are primarily genome browsers. In the following, we will present and discuss distinctive and unique features of UCNEbase.
UCNEbase is block-centric and provides complete synteny maps of UGRBs for many different vertebrate genomes, including orphan UCNEs located in unassembled contigs. With the exception of CONDOR, Ancora and TFCONES, all other resources do not assign conserved regions to GRBs.
Most other resources cover fewer species. For instance, cneViewer provides information for human and zebrafish only, TFCONES for human, mouse and fugu. UCNEbase covers 18 vertebrate genomes. Only Ancora and ECR browser offer data for a comparable number of species. Some existing resources are restricted to selected genomic regions. TFCONES considers only CNEs near transcription factor genes. The CONDOR database excludes elements outside synteny blocks.
With the exception of CONDOR and VISTA Enhancer browser, none of the other resources uses unique identifiers. CNEs are simply defined by genomic coordinates which will be outdated when a new genome assembly replaces the current one. UCNEbase is the only resource that uses informative names, indicating the evolutionary relationships between elements.
UCNEbase is highly interoperable with the UCSC genome browser. Most other resources display the data in their own browsers. Virtually all information from UCNEbase is available in custom tracks that are automatically pre-loaded by the hyperlinks to the USCS browser. This allows exploration of its content in a rich data environment.
DATABASE AVAILABILITY AND TECHNICAL SPECIFICATIONS
UCNEbase is publicly available at http://ccg.vital-it.ch/UCNEbase/ without need of preregistration. All data can be downloaded from the UCNEbase web site as flat files or as MySQL dumps, or by anonymous FTP from ftp://ccg.vital-it.ch/UCNEbase/.
UCNEbase is maintained as relational database using MySQL as database management system. The web interface was created with PHP and Java scripts and runs on an Apache web server hosted by the Vital-IT high-performance computing centre. The database schema diagram (ER model) is available from the UCNEbase web site.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods.
FUNDING
The Swiss National Science Foundation [PDFM33-120719 to S.D.]. Funding for open access charge: Swiss government.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Computationally intensive tasks were performed at the Vital-IT high-performance computing centre of the Swiss Institute of Bioinformatics.
REFERENCES
- 1.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 2.Dermitzakis ET, Reymond A, Lyle R, Scamuffa N, Ucla C, Deutsch S, Stevenson BJ, Flegel V, Bucher P, Jongeneel CV, et al. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature. 2002;420:578–582. doi: 10.1038/nature01251. [DOI] [PubMed] [Google Scholar]
- 3.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandelin A, Bailey P, Bruce S, Engstrom PG, Klos JM, Wasserman WW, Ericson J, Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24:344–352. doi: 10.1016/j.tig.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 6.de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 8.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 9.Woolfe A, Elgar G. Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv. Genet. 2008;61:307–338. doi: 10.1016/S0065-2660(07)00012-0. [DOI] [PubMed] [Google Scholar]
- 10.Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Retelska D, Beaudoing E, Notredame C, Jongeneel CV, Bucher P. Vertebrate conserved non-coding DNA regions have a high persistence length and a short persistence time. BMC Genomics. 2007;8:398. doi: 10.1186/1471-2164-8-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillier L, Miller W, Birney E, Warren W, Hardison R, Ponting C, Bork P, Burt D, Groenen M, Delany M, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 14.McEwen GK, Woolfe A, Goode D, Vavouri T, Callaway H, Elgar G. Ancient duplicated conserved noncoding elements in vertebrates: a genomic and functional analysis. Genome Res. 2006;16:451–465. doi: 10.1101/gr.4143406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson WR. Effective protein sequence comparison. Methods Enzymol. 1996;266:227–258. doi: 10.1016/s0076-6879(96)66017-0. [DOI] [PubMed] [Google Scholar]
- 16.Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 17.Akalin A, Fredman D, Arner E, Dong X, Bryne JC, Suzuki H, Daub CO, Hayashizaki Y, Lenhard B. Transcriptional features of genomic regulatory blocks. Genome Biol. 2009;10:R38. doi: 10.1186/gb-2009-10-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woolfe A, Goode DK, Cooke J, Callaway H, Smith S, Snell P, McEwen GK, Elgar G. CONDOR: a database resource of developmentally associated conserved non-coding elements. BMC Dev. Biol. 2007;7:100. doi: 10.1186/1471-213X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engstrom PG, Fredman D, Lenhard B. Ancora: a web resource for exploring highly conserved noncoding elements and their association with developmental regulatory genes. Genome Biol. 2008;9:R34. doi: 10.1186/gb-2008-9-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dieterich C, Wang H, Rateitschak K, Luz H, Vingron M. CORG: a database for COmparative Regulatory Genomics. Nucleic Acids Res. 2003;31:55–57. doi: 10.1093/nar/gkg007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persampieri J, Ritter DI, Lees D, Lehoczky J, Li Q, Guo S, Chuang JH. cneViewer: a database of conserved non-coding elements for studies of tissue-specific gene regulation. Bioinformatics. 2008;24:2418–2419. doi: 10.1093/bioinformatics/btn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee AP, Yang Y, Brenner S, Venkatesh B. TFCONES: a database of vertebrate transcription factor-encoding genes and their associated conserved noncoding elements. BMC Genomics. 2007;8:441. doi: 10.1186/1471-2164-8-441. [DOI] [PMC free article] [PubMed] [Google Scholar]