Abstract
The Rfam database (available via the website at http://rfam.sanger.ac.uk and through our mirror at http://rfam.janelia.org) is a collection of non-coding RNA families, primarily RNAs with a conserved RNA secondary structure, including both RNA genes and mRNA cis-regulatory elements. Each family is represented by a multiple sequence alignment, predicted secondary structure and covariance model. Here we discuss updates to the database in the latest release, Rfam 11.0, including the introduction of genome-based alignments for large families, the introduction of the Rfam Biomart as well as other user interface improvements. Rfam is available under the Creative Commons Zero license.
INTRODUCTION
The Rfam database categorizes non-coding RNAs and their conserved primary sequence and RNA secondary structure through the use of multiple sequence alignments (MSAs), consensus secondary structure annotation and covariance models (CMs) produced using the Infernal suite of software (1). Each family consists of a set of RNA sequences which are believed to share a common ancestor. We provide a representative alignment for the family (termed the seed alignment) that has been annotated with a consensus RNA secondary structure. A CM is built to describe the family, as well as a full alignment which represents all matches of the CM to sequences in our underlying sequence database. Our primary goal is to provide a comprehensive and accurate set of non-coding RNA annotations for genome annotation. Our alignments are also used extensively for training and benchmarking of other RNA sequence and structure analysis tools.
The procedure behind creating an Rfam family has been detailed in our previous publications (2). Briefly:
a MSA, known as the seed alignment, with secondary structure is produced by an Rfam curator or from a community submission;
a CM is created and calibrated from the seed using Infernal’s cmbuild and cmcalibrate programs;
the Rfam sequence database, Rfamseq, is searched with BLASTN using query sequences from the seed alignment. Surviving subsequences that pass an E-value threshold are then re-searched with the CM to define final hit locations and scores;
an Rfam expert curator sets a bit-score threshold, above which sequences are considered to be true homologues of the original seed sequences. Sequences with a bit-score below the threshold are not included in the family; and
the Rfam curator adds selected novel and diverse sequences resulting from the search to the seed alignment and the process is repeated until no new members of the family are found.
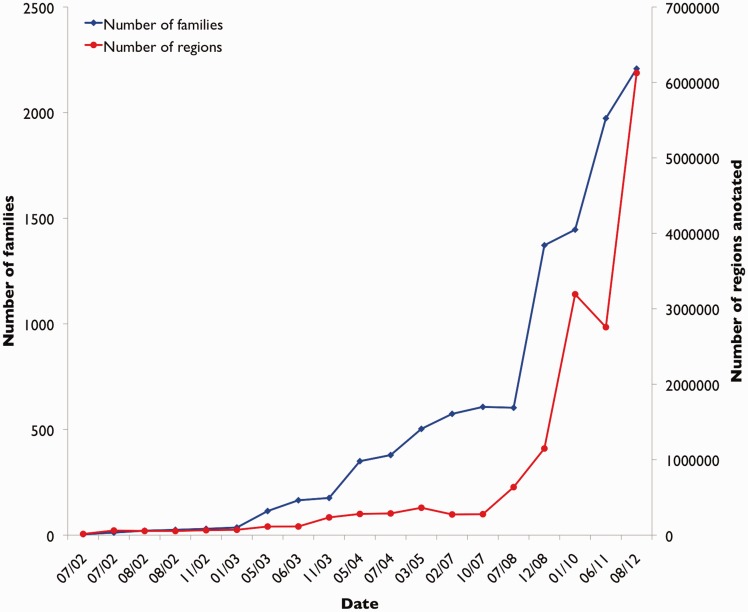
The first public release of Rfam, v1.0, was made available in July 2002. Rfam v11.0 represents the 10th anniversary of Rfam. As shown in Figure 1, the database has grown from 25 families in the first release to 2208 families in Rfam 11.0, and coverage has grown from around 50 000 sequence regions to over 6 million. Rfam is a large and comprehensive source of ncRNA annotation, covering all kingdoms of life and many types of functional non-coding RNAs. We have continued to improve our website and data access options, and now provide Biomart and RESTful interfaces. In addition to providing alignments and CMs, we provide cross-references to a number of other resources, such as links to PDB structures, ontology terms and cross-references to other relevant databases, such as the European Nucleotide Archive and mirBase. Our data are widely used in a number of different ways, such as vertebrate and bacterial genome annotation (3,4), as a dataset to the bioinformatics community for study in itself (e.g. identification of new family relationships through clustering), and as a controlled set of alignments for testing novel methodologies (5–7). In 2007, Rfam was one of the first biological databases to make its annotations available for editing in the online encyclopedia Wikipedia (8). This community annotation approach has proven to be highly successful in Rfam and in other successful Wikipedia annotation efforts, including our sister database Pfam (9) and the GeneWiki resource (10).
Figure 1.
Growth of Rfam families and sequence regions annotated per each Rfam release. Dates are in MM/YY format.
NEW DEVELOPMENTS
The Rfam 11.0 release
For Rfam 11.0, we have updated the underlying sequence database from that used for Rfam 10.1 and Rfam 10.0. Rfamseq 11 is based on the standard and whole-genome shotgun data classes from release 110 of the EMBL-Bank (January 2012). The previous version, Rfamseq 10, was based on the equivalent EMBL-Bank release from July 2009. In this time, the Rfam sequence database has grown by ∼34 million sequences; Rfamseq 11 is 1.6 times as large as Rfamseq 10 and contains 89 111 824 sequences.
As part of the sequence update, sequences belonging to the seed alignments are updated and all families are re-searched against the new sequence database. The increased sequence diversity has resulted in the majority of families increasing in size. Outside of the four largest families (tRNA, and the bacterial, eukaryotic and archaeal SSUs), the average family roughly doubled from its Rfam 10.1 size, in terms of numbers of sequences. The sequence update has enabled our families to identify more homologues; for instance the cspA thermoregulator family (RF01766) has grown from 434 sequences to 6179 sequences. This family now identifies actinobacteria, alpha- and beta-proteobacterial cspA thermoregulators, which were previously poorly represented in this family. Seventy-seven families decreased in size, primarily due to rethresholding to improve family specificity.
New families
Rfam 11.0 includes 246 new families and 4 new clans relative to Rfam 10.1. We added 2 extra subtypes to our family classifications: Gene;antitoxin (currently 5 families in this release) and Gene;lncRNA (225 families). We have also added families contributed by the community, such as rsmX (RF02144) and sRNA-Xcc1 (RF02221) which were submitted via the RNA Biology New Families track (11,12). We have continued to build families for long non-coding RNAs, and have added 144 such families, covering genes such as FTX, Sphinx and ZFAT. Due to the length of lncRNA genes, we do not attempt to build families to the entire length of a given long non-coding RNA; rather, we build several shorter families to regions of higher conservation within the lncRNA transcript. We have also added 52 new bacterial small RNA families and 11 new miRNAs.
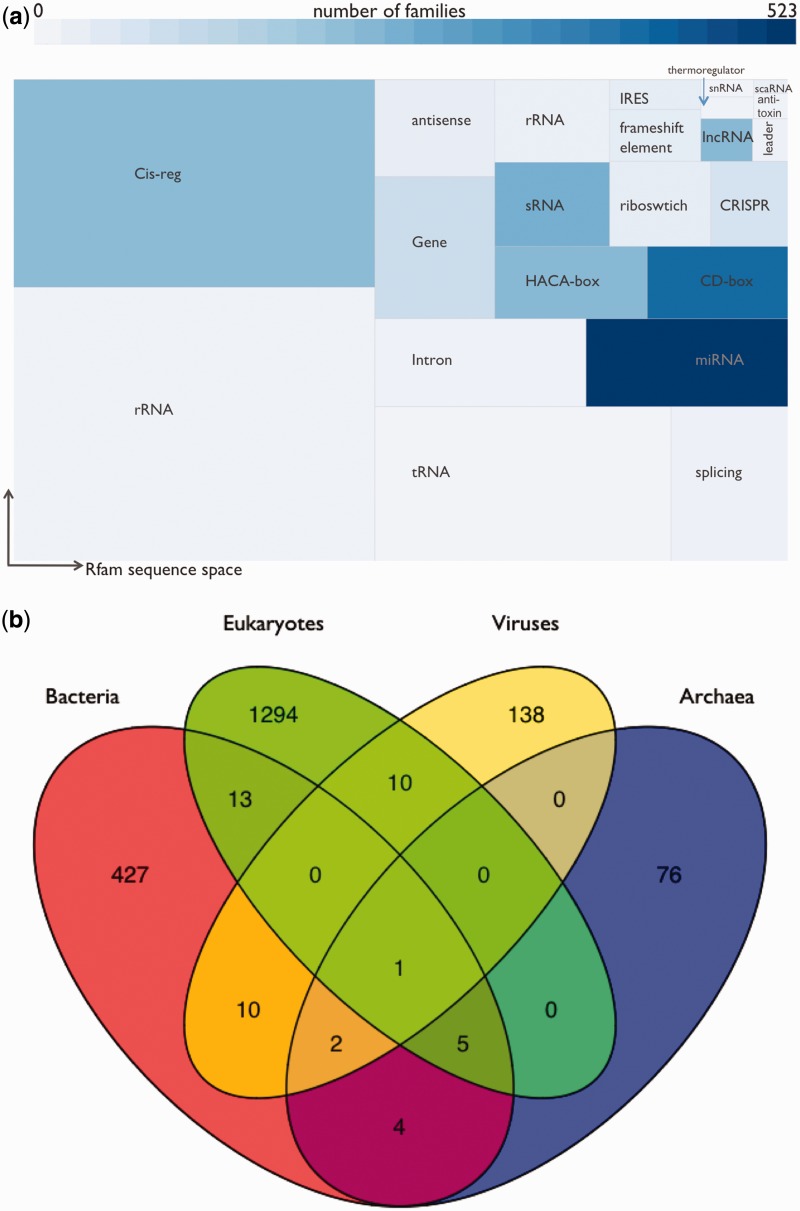
In Figure 2a, we illustrate the coverage of Rfamseq sequence space by Rfam family type, and in Figure 2b we present the taxonomic breakdown of Rfam families, based on the sequences present in the seed alignments. Families belonging to a clan have been treated as belonging to a single family; e.g. we treat the SSU families as a single large family where the seed contains eukaryotic, bacterial and archaeal members. Although there are only 6 rRNA families in Rfam, the ubiquity of these RNAs means that rRNAs are our single largest category of annotations, followed by the cis-regulatory elements (317 families). The largest categories of families are the miRNAs (523 families) and the snRNAs (678 families), yet they provide comparatively few sequence annotations. The large number of miRNA and snRNA families also skews the taxonomic distribution of Rfam families and means that the majority of Rfam families (1294) describe eukaryotic RNAs. Only the tRNA clan has seed sequences from all high-level taxonomic groups (viruses together with the three main kingdoms of life), while a further five families (RNase P, SRP, Group II catalytic introns, 5 S ribosomal RNA and TPP riboswitch) or clans have seed sequences from bacteria, archaea and eukaryotes, but are not known to occur in viruses. A full breakdown of taxonomic membership is presented in the Supplementary Material, and we refer the readers to Hoeppner et al. for a more comprehensive analysis of RNA family distribution throughout the kingdoms of life (13).
Figure 2.
(a) Rfam types organized according to their coverage of sequence space. Size of rectangles is proportional to the number of regions annotated by families of that Rfam type; colour of rectangles is proportional to the number of families of that type. (b) Taxonomic coverage of Rfam families. Families have been categorized according to the taxonomic groups covered by the seed sequences, and families in clans are treated as belonging to a single family. This analysis omits six families where the seed contains only unclassified sequences.
Towards genome-based alignments for Rfam families
The growth in the underlying sequence data has proved challenging for us to deal with for four families in particular: RF00005 (tRNA), RF00177, RF01959 and RF01960 (ribosomal SSU families). For these families, the alignments produced are extremely large, and in the case of the ribosomal RNA families the CMs are complex and computationally expensive to run. While the Infernal software is capable of building full alignments based on the entire Rfamseq database for these families, it is unfeasible to display these alignments and sequences on the website. We now provide two types of alignment for these families; the original full alignment (based on all sequence matches to Rfamseq), and an alignment based solely on sequences taken from completed genomes provided by the European Nucleotide Archive [http://www.ebi.ac.uk/genomes/]. The former is available from our FTP site, while the genome-based alignment is available through our website. The sizes of these alignments are given in Table 1. The smaller alignments, based on sequences from completed genomes, give family matches in a more biologically relevant context. This whole-genome alignment approach also allows us to better manage the growth in Rfam family alignments as the amount of sequence data available increases.
Table 1.
Comparison of alignment sizes for the four largest Rfam families between Rfam 10.1, full alignments and genome-based alignments
| Family | Description | Number of sequences in full, Rfam 10.1 | Number of sequences in full, Rfam 11.0 | Number of sequences in genome-based alignment |
|---|---|---|---|---|
| RF00005 | tRNA | 1 101 833 | 2 106 268 | 298 470 |
| RF00177 | Bacterial small subunit ribosomal RNA | 343 886 | 744 528 | 7429 |
| RF01959 | Archaeal small subunit ribosomal RNA | 9072 | 881 056 | 7394 |
| RF01960 | Eukaryotic small subunit ribosomal RNA | 45 117 | 65 901 | 425 |
Annotation of the RefSeq Collection
The NCBI Reference Sequence collection (RefSeq) is a non-redundant set of annotated sequences (14). As part of Rfam 11.0, we provide Rfam matches to the ncRNA section of RefSeq release 53. These annotations are available in a dedicated tab for each family, and are available on a per-organism basis in the Genomes section of the Rfam website. 860 Rfam families have matches to a RefSeq sequence, and we provide 21 283 annotations to 12 334 distinct RefSeq sequences. These annotations help our users link out from our families to externally curated sequences and relevant database cross-references and identifiers.
Wikipedia annotation of non-coding RNA families
The Rfam database is a pioneer in using Wikipedia for annotation of biological data (8). Since our Wikipedia annotation project started in 2007, we have linked to 923 pages of Wikipedia content, and the Rfam-Wikipedia model has been adopted by other biological databases, such as Pfam and miRBase (9,15). Both Pfam and Rfam use a common database to store information about Wikipedia editing activity. Since November 2011 for both Rfam and Pfam we have seen 15 548 edits to articles by 3721 Wikipedia editors. There are many different types of people who edit Wikipedia and we were interested to identify what sort of contributions different groups made. We were able to classify all editors into one of five types:
Scientists (92 editors)—editors that make scientific contributions to Wikipedia;
Rfam and Pfam scientists (12 editors)—editors who are, or have been, employed on the Pfam or Rfam projects;
Wikipedians (85 editors)—editors who make changes only to the non-scientific aspects of articles;
Robots (78 editors)—Editors that are automated scripts. They are characterized by having the word Bot in their Wikipedia user name; and
Long tail—(3454 editors) editors who had made a relatively small number of edits (<10) were grouped together. The long tail is actually a combination of scientists and Wikipedians, meaning the above numbers are underestimates.
We present a full breakdown of editors and their contributions (both addition and removal of text) in Table 2. Overall the editors in the long tail who made <10 edits each made the largest number of total edits to Wikipedia. This observation shows the importance of the contribution of occasional editors. The most prolific editors are the scientists involved in the Rfam and Pfam project. The 12 editors made 2604 edits and on average those editors have added 180 000 characters to the articles we link to. The Wikipedians on average delete more content than any other type of editor which reflects their role to keep the encyclopedia clear of vandalism and to uphold Wikipedia’s style guidelines. It is encouraging to see that the total amount of content added by scientists outside of the project is greater than that added by Xfam scientists.
Table 2.
Editing statistics for Wikipedia articles linked to by Pfam and Rfam classified by editor type. Character values rounded to nearest 1000
| Type | No. of edits | No. of editors | Total characters added (1000s) | Total characters subtracted (1000s) | Average characters added per Editor (1000s) | Average characters subtracted per Editor (1000s) |
|---|---|---|---|---|---|---|
| Xfam | 2604 | 12 | 2166 | 321 | 181 | 27 |
| Scientist | 955 | 92 | 2645 | 730 | 29 | 8 |
| Wikipedian | 2570 | 85 | 4033 | 2506 | 47 | 30 |
| Bot | 2783 | 78 | 730 | 337 | 9 | 4 |
| Long tail | 6636 | 3454 | 1767 | 402 | 1 | 0.1 |
| Total | 15 548 | 3721 | 11 340 | 4296 | 267 | 69 |
IMPROVEMENTS TO ACCESS
RESTful interface
We have added a ‘RESTful’ interface to many sections of the Rfam website. REST (or Representation State Transfer) is a convention for building websites that makes it easier to interact with the website programmatically. Such an Application Programming Interface (API) allows users to write scripts to retrieve data and access services, rather than having to use a browser or to ‘scrape’ pages to extract data. Most data can now be retrieved in a range of file formats, from JSON to XML. We also provide access to the single sequence search tool, allowing users to submit searches and retrieve results from a script or program. Full documentation for the RESTful interface can be found on our help pages at http://rfam.sanger.ac.uk/help#tabview=tab6, including sample Perl scripts that illustrate how it can be used.
The Rfam Biomart
A Biomart is a federated database system which aims to facilitate exploration and interrogation of large biological datasets (16). It has been adopted by many biological databases, such as Ensembl (17) and the Mouse Genome Database (18). Our aim in creating a Biomart was to make available sophisticated query technology in a user-friendly format; currently, the Rfam website only allows relatively simple querying by (e.g.), family accession, general search term, taxonomy or family type. Search parameters are referred to as filters, and the data requested are referred to as attributes. Available data filters and attributes are shown in Figure 3. The Biomart allows more complex queries involving a combination of search terms. For example, a user may obtain all the sequences annotated by Rfam as mouse miRNAs, or retrieve all Rfam families associated with a particular publication. The user is then able to examine the sequences resulting from their query, as well as ontology terms, bit scores and E-values. The first 1000 results of any query are returned on the website; if a search returns more than 1000 results, the full results are available for download using the links on the results page.
Figure 3.
Sunburst visualization of family taxonomy for RF01051, the cyclic di-GMP-I riboswitch. Users may select regions of taxonomic space and use the controls on the left to download an alignment of their chosen subset of species.
The Rfam Biomart is available through following the links on the Rfam website, or directly at http://xfam-biomart.sanger.ac.uk.
Sunburst and customized alignments for each family
Following the success of the sunburst species visualization in Pfam, we have adopted this same style of visualization for the Rfam website (Figure 3). In addition, users can use the sunburst to select a taxonomic subset of sequences for each family, and download an alignment of only those sequences in Stockholm file format.
UCSC Genome Browser tracks
Rfam data tracks are available for the organisms present in the UCSC Genome Browser and are available from our FTP site.
Future plans
As we have discussed previously, the relentless increase in the ENA means that it is increasingly impractical for us to base our full alignments on sequence data from the ENA. For instance, the SSU families’ full alignments now contain hundreds of thousands of sequences, and the large alignments are extremely computationally expensive to generate. Furthermore, these large alignments are difficult to store and manipulate. To address these issues, we have created alignments from sequences belonging to complete genomes only for five families, which have resulted in alignments of a much more manageable size. We hope that using sequences from completed genomes results in a much more useful and meaningful alignment. We intend to adopt a similar, genome-centric policy for all Rfam families in the future. Seed sequences will continue to be sourced from ENA to ensure that our seeds are as comprehensive as possible. If there is sufficient demand, we will also ensure that full annotations, based on the ENA standard and whole-genome shotgun data classes are available via FTP.
Infernal 1.1
A major update to Infernal, the software behind Rfam, was announced in July 2012 and will be used for the next Rfam release. (Infernal 1.1 was not available in time for work on Rfam 11.0.) The most significant improvement is a faster filter pipeline for database searches, based on the accelerated profile-HMM methods implemented in the HMMER3 software package (19), which makes searches roughly 100-fold faster than in the previous version. Infernal is now fast enough to obviate the need for the BLAST-based filtering scheme used by Rfam since its initial 1.0 release. However, the removal of the BLAST filters and other changes in Infernal will require that every Rfam family's thresholds be reexamined before the next release of the database. Importantly, the format of the CM files has changed slightly between v1.0.2 and v1.1 of Infernal; CM files for both versions are provided as part of Rfam 11.0.
Submit your alignment
We are constantly looking to improve existing Rfam alignments as well as to include new families. In order to make this as easy for our users as possible, we will soon be providing a web form for people to upload their Stockholm formatted alignments. These alignments will be checked by an Rfam curator before inclusion into an existing family or creation of a new family. We are also investigating methods by which we can roll out new families and improvements to existing families as swiftly as possible, to avoid the sometimes long periods between official Rfam releases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Dataset 1.
FUNDING
Wellcome Trust [WT098051 to S.W.B., J.D., R.E., J.T. and A.B.]; Howard Hughes Medical Institute (to E.P.N. and S.R.E.); Rutherford Discovery Fellowship administered by the Royal Society of New Zealand (to P.P.G.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully acknowledge the invaluable computational support of Pete Clapham, Guy Coates and David Harper of the Sanger Systems and Database groups, and of Goran Ceric of Janelia Farm’s High Performance Computing group.
REFERENCES
- 1.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato K, Kato Y, Hamada M, Akutsu T, Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27:i85–i93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer F, Kurtz S, Backofen R, Will S, Beckstette M. Structator: fast index-based search for RNA sequence-structure patterns. BMC Bioinformatics. 2011;12:214. doi: 10.1186/1471-2105-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 8.Daub J, Gardner PP, Tate J, Ramskold D, Manske M, Scott WG, Weinberg Z, Griffiths-Jones S, Bateman A. The RNA WikiProject: community annotation of RNA families. RNA. 2008;14:2462–2464. doi: 10.1261/rna.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Good BM, Clarke EL, de Alfaro L, Su AI. The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res. 2012;40:D1255–D1261. doi: 10.1093/nar/gkr925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen XL, Tang DJ, Jiang RP, He YQ, Jiang BL, Lu GT, Tang JL. sRNA-Xcc1, an integron-encoded transposon- and plasmid-transferred trans-acting sRNA, is under the positive control of the key virulence regulators HrpG and HrpX of Xanthomonas campestris pathovar campestris. RNA Biol. 2011;8:947–953. doi: 10.4161/rna.8.6.16690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moll S, Schneider DJ, Stodghill P, Myers CR, Cartinhour SW, Filiatrault MJ. Construction of an rsmX co-variance model and identification of five rsmX non-coding RNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol. 2010;7:508–516. doi: 10.4161/rna.7.5.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoeppner M, Gardner P, Poole AM. Comparative analysis of RNA families reveals distinct repertoires for each domain of life. PLoS Comput. Biol. 2012;8:e1002752. doi: 10.1371/journal.pcbi.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Haider S, Baran J, Cros A, Guberman JM, Hsu J, Liang Y, Yao L, Kasprzyk A. BioMart: a data federation framework for large collaborative projects. Database. 2011;2011:bar038. doi: 10.1093/database/bar038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]