Abstract
The ‘Standard European Vector Architecture’ database (SEVA-DB, http://seva.cnb.csic.es) was conceived as a user-friendly, web-based resource and a material clone repository to assist in the choice of optimal plasmid vectors for de-constructing and re-constructing complex prokaryotic phenotypes. The SEVA-DB adopts simple design concepts that facilitate the swapping of functional modules and the extension of genome engineering options to microorganisms beyond typical laboratory strains. Under the SEVA standard, every DNA portion of the plasmid vectors is minimized, edited for flaws in their sequence and/or functionality, and endowed with physical connectivity through three inter-segment insulators that are flanked by fixed, rare restriction sites. Such a scaffold enables the exchangeability of multiple origins of replication and diverse antibiotic selection markers to shape a frame for their further combination with a large variety of cargo modules that can be used for varied end-applications. The core collection of constructs that are available at the SEVA-DB has been produced as a starting point for the further expansion of the formatted vector platform. We argue that adoption of the SEVA format can become a shortcut to fill the phenomenal gap between the existing power of DNA synthesis and the actual engineering of predictable and efficacious bacteria.
INTRODUCTION
The failure of the early pioneers of recombinant DNA technologies to implement standards for the construction and nomenclature of plasmids (1) and other genetic tools has resulted in decades of chaotic development of cloning vectors and other molecular assets. While these circumstances have not limited laboratory studies, the necessity of fixed formats for vector organization and designation has become increasingly apparent as we enter the era of Systems and Synthetic Biology (2,3). We are witnessing a fast transition from the handling of a few-at-a-time genes toward the implementation of complex genetic circuits (4,5) and, ultimately, engineering living cells to acquire new-to-nature properties (6). However, the biological functions and devices that are necessary for such engineering may not be characterized to a point that makes them usable and predictable (2). Finally, the vast majority of genetic engineering efforts are made in Escherichia coli. This is an excellent host for the physical assembly of DNA parts into devices and modules (6), but, often, this organism is not the optimal platform for the deployment of the designed properties in a biotechnological setting, e.g. a bioreactor or released to the environment (7,8). Many attempts to expand the vector toolbox beyond enteric bacteria have been made in the past decades (9–12) and include a large suite of broad-host-range plasmids (13), transposons and transposon-vectors (14–16). The latter two methods have been instrumental for the random chromosomal insertion of mobile DNA (e.g. systems based on Tn5; 15,17,18), site-specific targeting of heterologous DNA sequences (Tn7 and CTX; 19) or a combination of the two (7,20). While such vectors have permitted the study and enhancement of many desirable properties of Gram-negative bacteria, their molecular architectures have been generally capricious and largely dependent on the immediate application for which they were created.
The regrettable lack of format of tools that are developed in different laboratories for the same purpose most often impede data comparison and typically require re-cloning of the same sequences into available familial vectors, which becomes a veritable bottleneck for engineering of devices that involve parts from various origins. Furthermore, the functional components of the vectors (e.g. origin of replication or selection determinant) are often assembled within the naturally occurring segments (which may carry undesirable internal sites) and pasted into existing restriction sequences. Frequently, broad-host-range vectors, which are useful for their specified purposes, end up being unnecessarily large and burdened with useless, if not fastidious, DNA sequences. While all of these issues may be ultimately resolved by the synthesis of entire genomes à la carte (21), the immediate future still needs better molecular tools for the following: (i) exploration, analysis and de-construction of the functional space of existing bacteria; (ii) re-construction of the system with the same or a different connectivity of constituents for creating new properties and (iii) deployment of the features of interest in the most advantageous carrier. In this background, it comes as a surprise that the vast literature on cloning vectors and genetic tools for bacteria has drifted toward extreme diversification rather than standardization and only a few instances can be noted that overcome this state of affairs. One example is the proposition of the BioBrick standard (22), which allows for the successive incorporation of DNA parts that are retrieved from a formatted repository of functional sequences to a cognate plasmid scaffold. This is a suitable platform for the recurrent physical assembly of DNA segments, but it does not tackle the issue of functionality or deployment of the constructs. A variant of the BioBrick approach is the BglBrick approach (23), which exploits the loss of BlgII and BamHI sites when DNA segments that are cleaved with either enzyme are ligated to each other in a fashion that allows for regeneration of a usable BlgII sequence after each round of insertion. This method has been useful for assembling a large collection of valuable vectors with various combinations of replication origins, selection markers and expression systems (24). Finally, one could entertain the assembly of plasmids à la carte by recombining relevant DNA sequences in vitro with suitable enzymatic cocktails (25,26). While all of these advances constitute steps in the right direction, they also ask for a breach from the more traditional cloning procedures, which may not fit the invested interests of a large community of existing users and available constructs.
The database (DB) described below is an attempt to overcome these problems by creating a coherent platform of molecular tools that are subject to a concise, minimalist and standardized format and nomenclature. Importantly, these tools are compatible with old and new cloning and DNA assembly methods. The resulting set of vectors, which are available in the DB, is composed of a number of synthetic, interchangeable and reusable functional modules that include broad-host-range origins of replication, antibiotic markers, expression systems and reporter genes that are punctuated by terminators and gadget-insertion sites. The first collection of these formatted vectors was intended for analysis and reassembly of engineered phenotypes in a suite of Gram-negative bacteria. We advocate that what we call the ‘Standard European Vector Architecture’ (SEVA) may become a fundamental reference to speed up the ease of biosystems engineering beyond laboratory applications.
DATABASE DESCRIPTION
Database organization
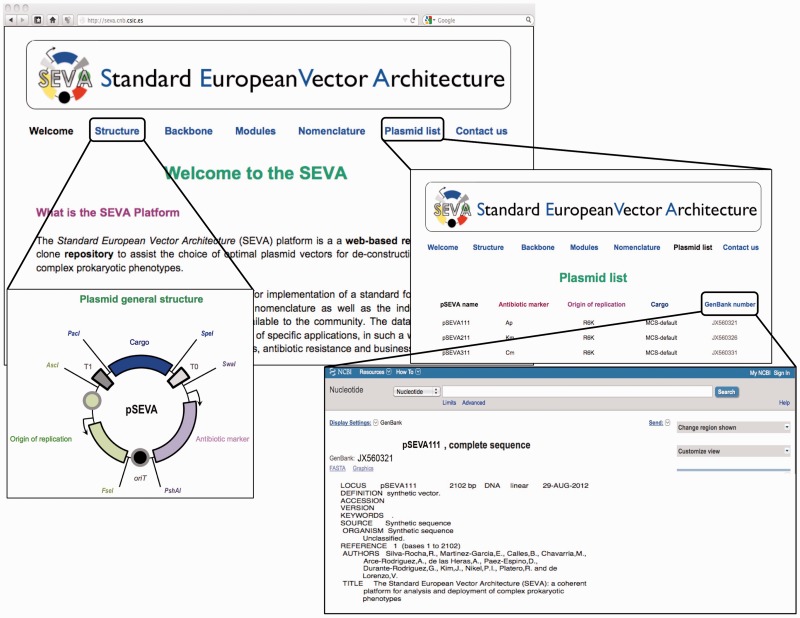
The SEVA-DB (http://seva.cnb.csic.es) is a resource for implementing a standard for the physical assembly of vector plasmids and their nomenclature. In addition, the DB serves as an index for the repository of functional sequences and the actual constructs that are available to the community. SEVA-DB was developed with an architecture that consists of a relational DB as the data storage layer, a series of modules that are hosted by an application server and a web-based presentation layer with an explicit set of standards that apply to all constructs. The DB was designed to simplify the choice of a given vector for specific applications in such a way that the user can easily decide what is the best configuration of replication origins, antibiotic resistances and business segments, as explained below (Figure 1). Furthermore, the user is given instructions on how to order vectors in the collection, contribute to the platform with new constructs, obtain a standard code for novel plasmids that follow the SEVA format and report problems. To this end, the tracking components include core data, clone-order data and user-management information. The core data include all relevant aspects of vector information such as origins of replication, antibiotic selection markers and cargo segments. The clones/vectors in the repository collection contain no inserts. Most clones are readily available; however, if mutations that are acquired during storage are detected, those clones may be temporarily removed from the active list of offered constructs, but their SEVA code and the link to their cognate DNA sequences to GenBank are maintained.
Figure 1.
Representative search results of the SEVA-DB. The welcome page includes a header with active links to the formatted plasmid structure, plasmid backbone, modules, nomenclature, plasmid list and contact information. Typically, the user will browse through the plasmid list, where the collection of constructs are ordered according to their origin of replication, antibiotic selection marker, type of cargo and a SEVA code. The page for each plasmid also has active links to the corresponding GenBank number and the complete DNA sequence of each construct.
The SEVA standard
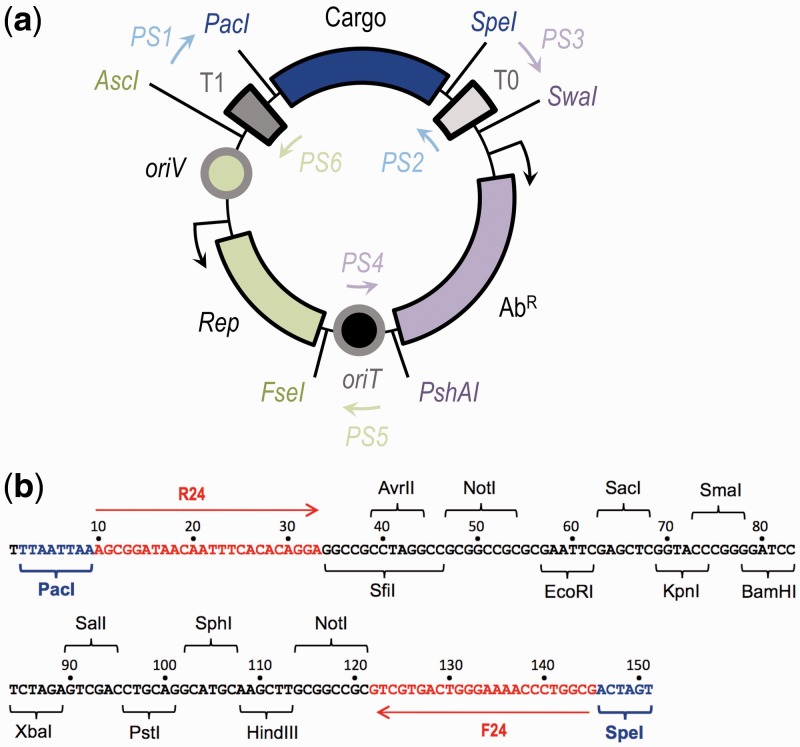
The SEVA platform involves the adoption of a plain set of rules for the physical assembly of the three basic components of plasmid vectors (i.e. origin of replication, selection marker and cargo segment, see below), edition and synthesis of the corresponding DNA sequences to remove any restriction sites that punctuate the general frame and implementation of an alphanumeric code for the resulting genetic tools. The schematic representation of the architecture of the SEVA vectors is shown in Figure 2a. To standardize the SEVA modules, a number of criteria were followed. First, each of the naturally occurring sequences that were destined for the various constructs were minimized to the shortest DNA segments that retained full functionality. The sequences were then erased of any of the restriction sites that were present in the polylinker of the classical vector pUC18 (27) and of SfiI, AvrII and NotI, but the encoded amino acids were retained by the mutated codons. The optimized fragments were produced by either site-directed mutagenesis of the DNA (28), as required, or were entirely chemical synthesized. Each of the three fragments was then assembled in a shared frame that was composed of three connecting parts. The plasmid vectors are, therefore, composed of six functional modules and the location and orientation of the three connectors provide the backbone of reference for the standardized vector collection. The sequences that link the variable parts include the strong, rho-independent transcriptional terminators T0 of phage lambda (103 bp) and T1 (105 bp) of the rrnB operon of E. coli (29). These terminators function by avoiding any transcriptional read-through into adjacent sequences, which increases plasmid stability (30). The third connecting element comprises a 246-bp DNA segment from the conjugative, broad-host-range plasmid RP4 (31) and is the plasmid’s very efficient origin of transfer (oriT). This oriT allows for conjugative mobilization of pSEVA plasmids into organisms in which no alternative transformation protocols are available (32; see below). Note that while oriT enables constructs to engage in conjugative gene transfer, its addition to SEVA vectors does not increase significantly chances of unintended uptake by naturally occurring microorganisms, which is largely caused by accidental DNA transformation. Because these three core sequences (T0, T1 and oriT) are shared by all plasmids of the series, specific primer pairs (PS1–PS6, Figure 2b) were designed that hybridize to these regions and can be used to amplify any of the intervening segments.
Figure 2.
Overall organization of structure of SEVA plasmids. (a) SEVA vectors are formed by three variable modules: a cargo (blue), a replication origin (green) and an antibiotic marker (magenta). Enzymes used to change the functional DNA segments are shown in the same color code and modules are separated by three permanent regions, which are shared by all vectors, the T0 and T1 transcriptional terminators and the oriT conjugation origin. All universal primers (PS1–PS6) are placed within the invariable backbone and are used to sequence/check the variable modules. (b) The structure of the default SEVA cargo. Cargos are cloned as PacI/SpeI fragments. The default segment contains the typical pUC18 polylinker enzymes from EcoRI to HindIII (the completely ordered restriction enzyme list is highlighted in the figure). Additional enzymes (i.e. SfiI, AvrII and NotI) are placed outside of the polylinker for specific cloning purposes. Within the cargo sequence, the enzyme recognition sites and the hybridization site of the universal M13 (R24/F24) primers are shown.
The first variable part of the pSEVA constructs consists of the antibiotic selection marker. As shown in Figure 2a, the DNA segments that carry such markers include the structural gene for one antibiotic (Ab) resistance gene and its native promoter. The unusual SwaI and PshAI restriction sites flank the Ab resistance unit and the expression of the Ab gene is oriented toward the oriT-connecting module. The size of the DNA segment for such an Ab resistance unit ranges from 0.8 to 1.3 kb, depending on the specific marker (see below). The second exchangeable part of the SEVA vectors is a DNA segment that holds the origin of replication of the plasmid, which is produced and inserted in the plasmid frame as an AscI-FseI fragment. The composition of such segment that endow replication is more complex than the one-gene counterparts for antibiotic selection because they include an oriV for replication initiation and may encode specific replication proteins and their corresponding promoters. Although the orientation of these sub-components may vary, the SEVA standard entails that the oriV sequence be proximal to the AscI end of the DNA fragment and that transcription of the replication protein(s), if any, points toward the FseI site of the oriT connector (Figure 2a). The broad-host-range replication origins that are selected to this end have different sizes (1.6–3.7 kb) and endow the plasmids with different copy numbers, which are decided by the user. Finally, the third variable element of the SEVA constructs is the DNA portion that bears the main functionality of the vector. We have designated this element as the ‘cargo module’ (31) and this module is always formatted as a PacI-SpeI fragment. This region confers a specific purpose to the plasmid, whether for cloning, expression of heterologous genes, creating reporter gene fusions or for chromosomal integration. Note that DNA fragments that are bordered by PacI-SpeI sites can also be cloned into the synthetic transposon vector of pBAM1 (31,33), thus allowing for stable chromosomal integration of a DNA segment that is assembled in SEVA plasmids. Figure 2b shows the sequence of the default SEVA cargo, which consists of a basic polylinker that is formed by the array of unique cloning sites of pUC18 (27), SfiI/AvrII and NotI sites upstream of the EcoRI site and a second NotI site that is placed downstream of HindIII site. These flanking NotI sequences allow concatenation of cloned fragments (positioned between EcoRI and HindIII) into derived SEVA vectors where NotI is unique (see below). This format increases the number of fragments that can be assembled into a SEVA cargo. The same principle is used for the SfiI site (18), but note that the AvrII site is used with PacI to implement various expression systems or other genetic devices, as explained in the next sections. The M13 phage F24 and R24 primer sequences are present in the default cloning cargo to allow for verification of inserted fragments. Figure 2b also highlights the restriction site sequences and the positions of the M13 primers.
Nomenclature of the pSEVA plasmids
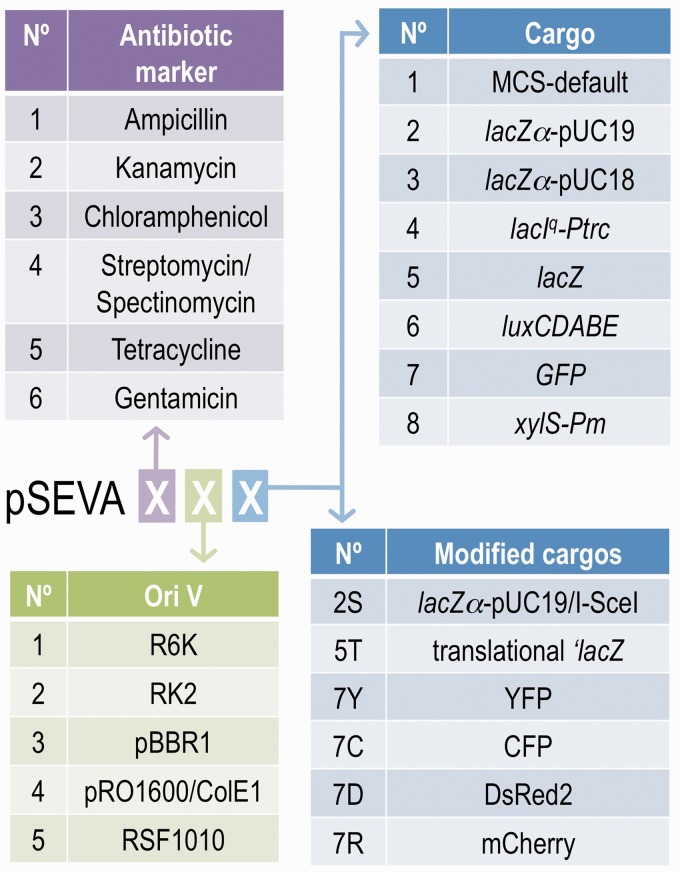
In addition to the standardized layout of the constructs that is sketched in Figure 2a, the SEVA format calls for the designation of each of the vectors with the code that is summarized in Figure 3. Under this criterion, all vectors are named pSEVA followed by a multi-digit cipher. The first position of the cipher reflects the antibiotic resistance marker, and at the time of writing this paper, six had been used (see below). The second position of the code is for the origin of replication (five types thus far). Finally, the third spot indicates the more variable cargo module (eight types for now). Because a large number of cargos are possible, the third position can be extended with serial numbers and further qualified by using one or more upper/lower case Latin letters that act as a prearranged ID code. Finally, vectors with a genetic gadget that is inserted at the unique sites between the functional modules and the connector sequences are to be noted with a Greek letter that is placed before or after the position corresponding to the more proximal segment. The default pSEVA111 vector from which all others originate has an ApR marker gene, the narrow-host-range origin of replication R6K and the default SEVA polylinker, as explained in the next sections.
Figure 3.
SEVA nomenclature. Plasmid vectors are named using digits that reflect their functional modules. The codes include at least three positions: the first stands for antibiotic markers, the second for the replication origin and the third for the cargo. The lists of possible modules for each position are given, different versions of cargos are possible, and additional digits and letters are used to specify the modification (as shown in the lower right list).
The pSEVA housekeeping components: antibiotic resistances and origins of replication
The first series of standardized antibiotic markers comprise those that are commonly used for selection in Gram-negative bacteria. The set of generated cassettes included genes for resistance to ampicillin (Ap, acquired from the bla gene of pBAM1; 31), kanamycin (Km, the aphA gene was amplified from the pBAM1 plasmid; 31), chloramphenicol (Cm, the cat gene from the pCC1FOS plasmid; Epicentre Biotech), streptomycin/spectinomycin (Sm/Sp: the aadA gene from pVLT35 plasmid; 10), tetracycline (Tc, the tetA gene from plasmid pBBR1-MCS3; 12) and gentamicin (Gm, the aacC1 gene from pBBR1MCS-5; 12). A complete description of the ‘wet’ standardization process that was performed for each exchangeable module will be published elsewhere (E. Martínez-García et al., manuscript in preparation).
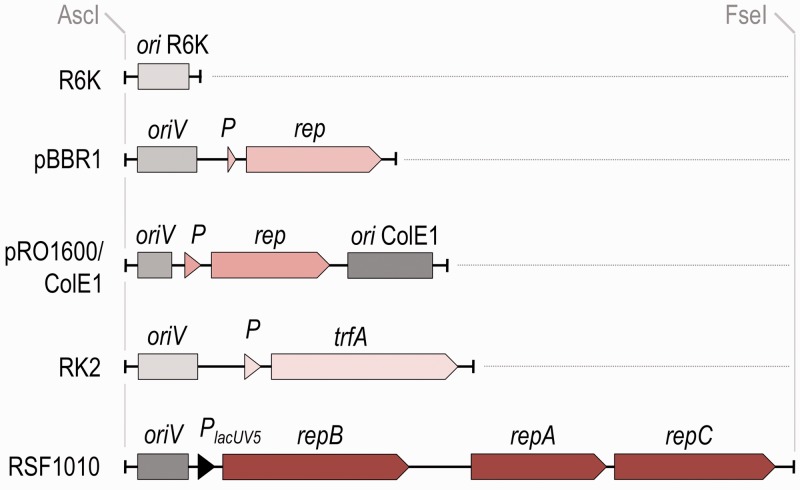
The antibiotic markers were then combined into the SEVA frame in Figure 2a with a variety of origins of replication, i.e. narrow-host range and broad-host-range (Figure 4). In this article, we have focused on origins that are more commonly used and can be employed for manipulations of environmental, Gram-negative bacteria. One extreme of the series is the narrow-host range R6K origin. Replication of the R6K origin is dependent on the Π protein (34); therefore, it can only be maintained in E. coli cells that express the pir gene in trans (35). This characteristic makes R6K very useful for genome-editing techniques, where chromosomal integration of a specific plasmid is required (31,36). The standardized oriV R6K includes a minimal 392-bp sequence that was excised from pBAM1 (31) and combined with the default cargo and each of the antibiotic markers. The resulting R6K-vectors were then used as templates for replacing the replication module with a number of broad-host-range (BHR) counterparts that were edited as indicated previously and formatted in all cases as an AscI-FseI fragment (Figure 4). The architecture of these origins consists of a minimum sequence for replication initiation (oriV) and one or more replication proteins (rep genes). The collection of reference constructs includes four of these (see sketches of Figure 4). The first and smallest (1.52 kb) BHR origin comes from the medium-copy number and small broad-host-range plasmid pBBR1, which was first isolated from the Gram-negative bacterium Bordetella bronchiseptica (37,38). This module is the basis of a large number of vectors (12,39) and includes only an oriV and a replication protein. One downside of such a DNA segment is that its copy number may be disadvantageous for some applications, e.g. regulation studies, but it is an excellent and stable replicon for most purposes. The next-larger module contains a hybrid of the origins of the narrow-host-range E. coli plasmid ColE1 (27) and the replication determinants of pRO1600, which is plasmid from a Pseudomonas aeruginosa isolate whose functionality is limited to closely related species (40). The ColE1 and pRO1600 origin combination is derived from vector pUCP24 (11), which was used to produce a 1.97-kb replication module for the SEVA collection. Note that a plasmid with the ColE1 oriV is found at a high copy number in E. coli, while the number of plasmids with the pRO1600 origin may vary from host to host (41). The third origin is the 2.22-kb replication origin of the broad-host-range IncPa plasmid RK2 (42). The standardized ori segment is formed by oriV followed by the gene that encodes the replication protein TrfA, which is expressed from its native BHR promoter (Figure 4). The immediate source of this RK2 origin was pJB785TT (43) and the relevant sequence of this origin was used for formatting the corresponding module. This origin of replication is among the least restrained and it keeps its copy number per cell very low (44). Finally, we formatted and edited a minimized version of the replication determinant of the IncQ, hyper-promiscuous replicon RSF1010 (45). The host range of the IncQ plasmids includes the enterics, pseudomonads and their proteobacteria relatives and more distant species (i.e. firmicutes, actinomycetes, cyanobacteria and acidophilic bacteria that are associated with mining environments). Not surprisingly, RSF1010 has been extensively exploited since the onset of genetic engineering for developing a large number of plasmid vectors (10,46). However, the extant arrangement of the RSF1010 replication elements were not optimal for our purposes because the oriV and the cognate replication proteins are encoded in DNA segments that are intertwined with mobilization and transfer determinants (45). To overcome this shortcoming, we created a streamlined segment that only contained the RSF1010 oriV and was positioned adjacent to the repBAC genes, which were expressed through an engineered PlacUV5 promoter (Figure 4; 47). The resulting synthetic origin is only 3.7 kb, which is smaller than the 5.4-kb region of the naturally occurring replicon (48,49). This new origin splits the replication of plasmids containing this module (as well as any of the previously mentioned origins of replication) from their conjugal transfer abilities. This property is present by default in all SEVA constructs by virtue of the third FseI-PshAI connector element of the plasmid backbone (Figure 1a), which bears the oriT of the BHR and self-transmissible RK2/RP4 plasmid that was mentioned earlier. Note that the conjugation machinery of this plasmid is one of the most promiscuous because it can deliver DNA to other Gram-negative and Gram-positive species and even to yeast (50) and mammalian cells (51). Furthermore, efficient mobilization of RK2oriT-containing plasmids can be brought about by transient expression of the corresponding tra/mob genes (32,52), which is a property that has been exploited for a long time to deliver exogenous DNA to hosts that are recalcitrant to straight transformation (53).
Figure 4.
Organization of the edited SEVA plasmid replication origins. The R6K suicide origin lacks the gene encoding the Π replication protein, which makes its maintenance dependent on the trans delivery of that protein. RK2 is formed by the vegetative origin (oriV) and the replication protein trfA. pBBR1 has a similar structure to RK2; however, the pRO1600/ColE1 origin is a hybrid of the pRO1600 origin and the ColE1 replication sequence. For RSF1010, the mob genes have been eliminated and the synthetic sequence has only the oriV and the repBAC genes, which are expressed from a PlacUV5 promoter.
The six antibiotic resistance markers were combined with the five, above-mentioned origins of replication in the vector frame depicted in Figure 2a that bears the default polylinker in Figure 2b. Doing so created a collection of 30 reference vectors, which are listed and described in the SEVA-DB. Notice that their different replication machineries are unlike each other, thereby ensuring their compatibility in the same bacterial host if they are co-selected with the appropriate antibiotics. Notice also that the plasmids are neither endowed with specific replication termination signals (54) nor with active segregation systems (55), which are specific for each plasmid and host in their natural settings. In the absence of these functions, cells containing SEVA plasmids are expected to resolve replication intermediates through the action of housekeeping topoisomerases (56) and to maintain the constructs through antibiotic selection. If this becomes a problem in specific hosts, the vectors can be improved for a given purpose by inserting toxin–antitoxin or counter-segregation systems (e.g. a parB cassette; 57) at the unique ‘gadget sites’ that were purposefully left available in the vector frame at the boundaries between the functional modules and the connecting elements (Figure 2). Note that the compatibility of different but compatible SEVA plasmids that encode diverse functionalities in the same cell expands the number and range of possible applications of these vectors.
Cargos for cloning and genome editing
The ‘cargo module’ is the most important part of the SEVA vectors because it is where the user implants the sequences to be examined (e.g. regulators, promoters and other actuators). A number of different cargos have been initially set up for the starting vector collection. The SEVA standard states that the main transcription flow of whatever device is designed as a cargo should proceed from the PacI site toward the SpeI site of the plasmid frame (Figure 2a). In this way, the intervening segment is transcriptionally insulated upstream by the T1 terminator, which is present in the first connecting sequence and downstream by the T0 terminator. The repository of produced cargos includes four types of applications. First, ‘cloning cargos’ include (in addition to the default polylinker of the vector in Figure 2b) a SEVA-formatted variant of the widely used lacZα cloning platform for blue/white screening of transformed cells containing plasmids with the cloned fragments on X-gal plates (27). These cargos also afford the expression of heterologous genes from the endogenous Plac promoter when placed in the correct orientation. As shown the SEVA-DB, the pUC19/18 cloning cargos were combined with the pBBR1 and pRO1600/ColE1 origins of replication and all possible antibiotic markers, thus generating 24 BHR cloning vectors. A special collection of lacZα-containing plasmids with the narrow-host-range oriV R6K were separately developed for editing (i.e. inserting into, deleting portions of or replacing specific portions of) the genomes of Gram-negative bacteria (31). In this case, the cargo consisted of the lacZα polylinker of pUC19, which was flanked in-frame by two I-SceI-targeted sequences for site-specific genomic deletions. Such a formatting (and the resulting six additional vectors, see SEVA-DB) expands the utility of the original procedure (36) to bacteria that are naturally resistant to Km, which was the only selection marker that was available up to that point for the genome-editing method.
Broad-host-range heterologous expression
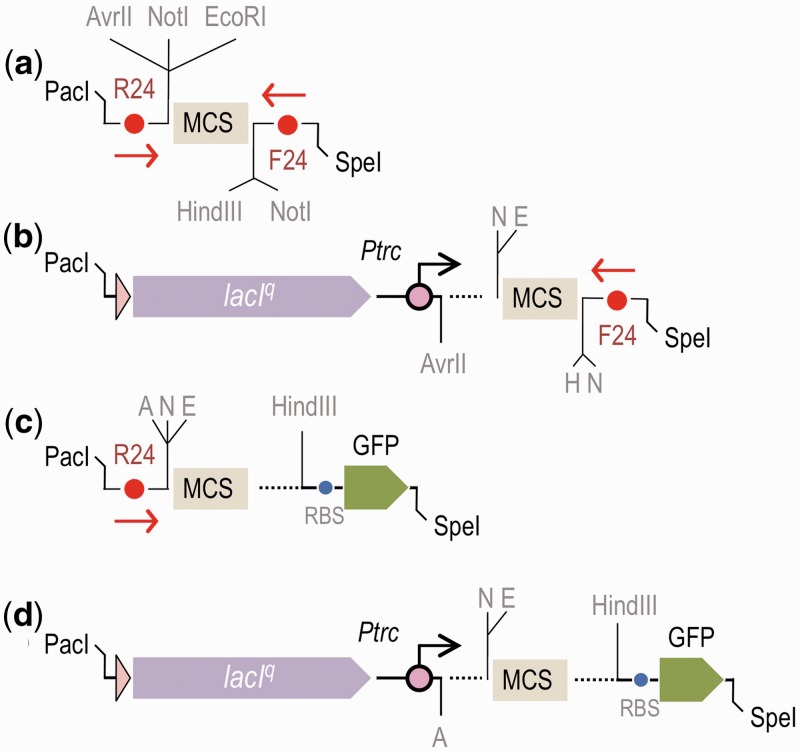
Other types of cargos were designed for the regulated expression of heterologous genes. Typical genetic devices to this end include one transcription factor (TF), the activity of which is triggered or inhibited by a specific inducer and a target promoter, upon which the regulator drives transcription of the desired gene(s). Two expression systems of this sort were formatted following the SEVA standard as part of the reference plasmid collection. The first system consists of plasmids containing the IPTG-inducible lacIq-Ptrc system, which was excised from the pTrc99a vector (58) and assembled in the SEVA frame as a PacI-AvrII fragment so that the regulated promoter points toward the AvrII site. In this way, users can benefit from the remaining downstream cloning sites of the default polylinker (Figure 2b). Note also that this expression cargo (as well as any other SEVA counterpart, see below) lacks translation signals (e.g. Shine-Dargarno ribosome binding sequences), which may need to be engineered on a case-by-case basis (59,60). A second broad-host-range expression system includes the benzoate/m-toluate-responsive XylS regulator of the TOL plasmid of Pseudomonas putida mt-2 and its cognate promoter Pm. The system is configured like the pJP plasmid series (9), but the DNA was edited so that five restriction sites that were found in the original sequence were removed and the segment was generated as a PacI-AvrII restriction fragment. The SEVA-DB displays a non-exhaustive list of pSEVA variants with each of the two expression cargos, which are merged with the R6K, RK2 or pBBR1 origins of replication and different antibiotic markers. The combination of the two expression modules with various plasmid copy numbers cover most transcriptional capacities and dynamic ranges (see below) that are usually required, but they can be easily replaced by other expression cargos and expression devices to satisfy particular specifications. One large source of regulatory cargos is the many TFs that regulate the expression of biodegradative operons that encode xenobiotic and recalcitrant compounds in soil bacteria (61,62).
Parameterization of promoter output
The last type of cargo that is available in the basic SEVA collection consists of a compilation of reporter genes for the visualization and quantitative measurement of expression in time and space. Reporters allow for the parameterization of regulatory interactions (e.g. the characterization of the transfer function of a given regulator-promoter pair). In most cases, the reporter product is encoded within a promoterless cassette, which contains the corresponding ORF and is preceded by a RBS. When a reporter is placed downstream of a transcription initiation device (e.g. a regulated promoter), its readout reflects the intensity of the transcriptional output. In other cases, the coding sequence of the reporter product is fused upstream or downstream of another gene to generate a translational fusion. Under these conditions, the optical or enzymatic readout of the hybrid reports its transcription and translation. It follows that when transcription is constant, translational fusions report the actual production of the hybrid protein. These features have a large number of applications for characterizing riboswitches (63) and small RNAs (64). In this background, we have formatted some of the most common reporters as HindIII/SpeI fragments so that they can be easily inserted downstream of any transcription initiation device at the default SEVA polylinker. The reporter cargos include intact and truncated versions of the β-galactosidase gene of E. coli (lacZ, ∼3 kb), which can be fused to another gene to make either transcriptional or translational fusions to this widespread indicator gene (65). This reporter is the only thus far whose activity can be expressed in absolute units (i.e. Miller units) and, despite its shortcomings (e.g. lengthy manipulations of cells, lack of an internal standard of reference; 66), it is still a very useful asset for studies on promoters in virtually any biological host. Similarly, the collection also contains the complete 5-kb, promoterless luxCDABE operon from Photorhabdus luminescens that encodes all enzymatic activities for emission of light (16). This reporter can be followed without any disruption of the cells and quantified very accurately (65). The reaction that releases photons is dependent on the physiological status of cells, specifically on FMNH2, which is a cofactor that sharply declines as cells enter stationary phase (67). Still, the lux reporter is optimal for measuring promoter output in steady-state growing cells, which is a common circumstance in laboratory experiments. Finally, the reporter cargo collection has five promoterless genes that encode different fluorescent proteins (i.e. GFP, CFP, YFP, dsRed2 and mCherry), all of which are formatted following the HindIII → SpeI standard of the SEVA frame. Because fluorescence is not diffusible, these reporters provide extraordinary detail, such as stochasticity, phenotypic variation and transcriptional noise, on gene expression at the single-cell level (68,69). Furthermore, the same bacteria can be engineered to express fluorescent proteins of various colors that are borne by either the same vector or by compatible SEVA plasmids, thereby allowing the monitoring of different processes in individual cells. Note that each of the reporter genes of the SEVA-DB has been standardized to also have the same, optimized RBS sequence (AGGAGGAAAAACAT) that was described for GFP of the pGreenTIR vector (70). The maintenance of the same architecture in the reporter cargos allows for comparison of results that are generated using different indicator genes and for the production, whenever needed, of conversion tables for different units of measurement. Ultimately, we expect these constructs to become instrumental for assigning the values of reporter readouts to absolute counts of RNA polymerase per second (PoPS counts; 65) of the regulatory node that is under investigation. Following the above-described numbering procedure, cognate plasmids are assigned the numeral 7 in the third position of the SEVA code because they are all variants of the original GFP protein, but the numeral is then followed by a letter to indicate the cargo: 7 for GFP, 7Y for YFP, 7C for CFP, 7D for DsRed2 and 7R for mCherry.
Sharing and distribution of SEVA plasmids
Plasmids that are available in the SEVA-DB and those that will enrich the collection in the future are distributed worldwide to researchers at any academic or non-profit laboratory. No MTAs will be requested prior to shipping of the constructs to potential users of such categories. When utilizing the constructs for commercial or industrial purposes, solicitors should be aware that some of the DNA sequences that were included in the SEVA plasmids could be subject to intellectual property restrictions. In this regard, the SEVA platform disclaims any carrier liability. Contributors to the collection must adhere to such an open-access policy or they should not number their constructions with the SEVA nomenclature. Clones are distributed either as glycerol stocks, stabs in agar tubes or purified plasmid DNA and in keeping with accepted practices (71), the SEVA-DB will distribute petitions of a small number of clones at no cost to the user but may charge handling, maintenance and shipping fees for large requests.
Users are encouraged to produce their own cargoes and other segments in the format that is advocated in this article and communicate those sequences to the plasmid repository through the SEVA-DB web page for quality control. As mentioned earlier, each of the SEVA constructs is allocated a core, multi-digit code that ciphers its physical organization. This code can be enriched with Latin and Greek letters, which consign variants of the default modules or genetic gadgets, to endow plasmids with specific properties. Contributions to the collection of SEVA vectors must be accredited by the curators of the platform to follow the above-specified standards, stored in the SEVA repository and assigned a definitive code prior to public release. Furthermore, users are discouraged from assigning SEVA codes to constructs that are not explicitly endorsed by the SEVA-DB managers.
FUTURE PERSPECTIVES
The structure of the SEVA plasmids allows for the straightforward combination of expression systems and reporter (or otherwise heterologous) genes such that any input (e.g. IPTG or benzoate) can be genetically wired to any output (lacZ, lux, GFPs) and the plasmid will still retain the intact multiple cloning site (from EcoRI to HindIII) that can be used for introducing other combinations of TFs and promoters à la carte (the possible ways of connecting SEVA cargos are exemplified in Figure 5). This modularity and the compatibility of the various replicons allow for the assembly of complex circuits in the same host and the performance/behavior of each sub-circuit to be easily monitored. Still, we argue that the importance of the SEVA platform is the expansion of the contemporary Synthetic Biology toolbox for bacteria to actual applications, e.g. for biocatalysis or bioremediation (72–75). To this end, the current SEVA-DB focuses on a number of broad-host-range replication origins and we have left typical E. coli replicons aside (22).
Figure 5.
Combination of expression and reporter cargos in the SEVA plasmids. (a) The structure of the default cloning cargo with the universal M13 primers (F24 and R24) and relevant restriction sites. (b) Assembling expression systems as PacI/AvrII fragments removes the R24 primer sequence and SfiI sites upstream of the AvrII site. (c) Cloning of reporter genes as HindIII/SpeI fragments removes the F24 primer sequence and a NotI site that is placed downstream of the HindIII site. (d) Combination of an expression system and reporter gene leaves an intact polylinker (from EcoRI to HindIII) and a unique NotI site between the AvrII and EcoRI sequences, which can be used to clone large constructs.
Supplementary Table S1 shows a non-exhaustive list of organisms where the current list of SEVA vectors have been tested and found to be functional (13). The RK2- and RSF1010-based vectors are, as expected, the most promiscuous. However, it is likely that the host range of the pBBR1-based and pRO1600-based vectors are understimated because they have been tried in only a small number of hosts. The modularity of the SEVA format allows for growth of the collection toward other replicons, such as those containing functional parts of Gram-positive organisms. The same holds true for the other functional segments, in particular, the cargo modules. Cargos are being expanded in our laboratory to include genetic mobile elements, new regulated-expression devices, systems for detecting/monitoring protein–protein interactions and various others. The community is encouraged to contribute to this effort to enrich the SEVA collection. New constructs will be entered in the SEVA-DB and made available as soon as they are verified to follow the standard. In summary, we advocate that adoption of the SEVA format will multiply the possibilities of programming a large number of bacteria beyond E. coli and will thus help to ease the transition between trial-and-error genetic engineering to systems-based Synthetic Biology (6,65,76).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
Funding for open access charge: BIO and FEDER CONSOLIDER-INGENIO Programs of the Spanish Ministry of Economy and Competitiveness, the MICROME and ST-FLOW Contracts of the EU, the ERANET-IB Program and funding from the Autonomous Community of Madrid (PROMPT).
Conflict of interest statement. None declared.
REFERENCES
- 1.Novick RP, Clowes RC, Cohen SN, Curtiss R, III, Datta N, Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol. Rev. 1976;40:168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 3.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 4.Michalodimitrakis K, Isalan M. Engineering prokaryotic gene circuits. FEMS Microbiol. Rev. 2009;33:27–37. doi: 10.1111/j.1574-6976.2008.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 6.Voigt CA. Genetic parts to program bacteria. Curr. Opin. Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 7.de Las Heras A, Carreño CA, de Lorenzo V. Stable implantation of orthogonal sensor circuits in Gram-negative bacteria for environmental release. Environ. Microbiol. 2008;10:3305–3316. doi: 10.1111/j.1462-2920.2008.01722.x. [DOI] [PubMed] [Google Scholar]
- 8.Cases I, de Lorenzo V. Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int. Microbiol. 2005;8:213–222. [PubMed] [Google Scholar]
- 9.Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in Gram-negative bacteria. Plasmid. 1997;38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Eltis L, Kessler B, Timmis KN. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 11.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 12.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 13.Lale R, Brautaset T, Valla S. Broad-host-range plasmid vectors for gene expression in bacteria. Methods Mol. Biol. 2011;765:327–343. doi: 10.1007/978-1-61779-197-0_19. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Herrero M, Sánchez JM, Timmis KN. Mini-transposons in microbial ecology and environmental biotechnology. FEMS Microbiol. Ecol. 1998;27:211–224. [Google Scholar]
- 15.de Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 16.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 17.Herrero M, de Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J. Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweizer HP. Vectors to express foreign genes and techniques to monitor gene expression in Pseudomonads. Curr. Opin. Biotechnol. 2001;12:439–445. doi: 10.1016/s0958-1669(00)00242-1. [DOI] [PubMed] [Google Scholar]
- 20.Silva-Rocha R, de Lorenzo V. A GFP-lacZ bicistronic reporter system for promoter analysis in environmental Gram-negative bacteria. PLoS One. 2012;7:e34675. doi: 10.1371/journal.pone.0034675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 22.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, Keasling JD. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 2010;4:1. doi: 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nour-Eldin HH, Hansen BG, Norholm MH, Jensen JK, Halkier BA. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006;34:e122. doi: 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 28.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stueber D, Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1:1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Garcia E, Calles B, Arevalo-Rodriguez M, de Lorenzo V. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 2011;11:38. doi: 10.1186/1471-2180-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JH. A Short Course in Bacterial Genetics. Harbor, NY: Cold Spring; 1992. [Google Scholar]
- 33.Martinez-Garcia E, de Lorenzo V. Transposon-based and plasmid-based genetic tools for editing genomes of Gram-negative bacteria. Methods Mol. Biol. 2012;813:267–283. doi: 10.1007/978-1-61779-412-4_16. [DOI] [PubMed] [Google Scholar]
- 34.Kolter R, Inuzuka M, Helinski DR. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 35.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Garcia E, de Lorenzo V. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 2011;13:2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x. [DOI] [PubMed] [Google Scholar]
- 37.Antoine R, Locht C. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from Gram-positive organisms. Mol. Microbiol. 1992;6:1785–1799. doi: 10.1111/j.1365-2958.1992.tb01351.x. [DOI] [PubMed] [Google Scholar]
- 38.Szpirer CY, Faelen M, Couturier M. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 2001;183:2101–2110. doi: 10.1128/JB.183.6.2101-2110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller WG, Leveau JH, Lindow SE. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 40.Olsen RH, DeBusscher G, McCombie WR. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J. Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farinha MA, Kropinski AM. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl Acad. Sci. USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santos PM, Di Bartolo I, Blatny JM, Zennaro E, Valla S. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 2001;195:91–96. doi: 10.1111/j.1574-6968.2001.tb10503.x. [DOI] [PubMed] [Google Scholar]
- 44.Thomas CM, Cross MA, Hussain AA, Smith CA. Analysis of copy number control elements in the region of the vegetative replication origin of the broad host range plasmid RK2. EMBO J. 1984;3:57–63. doi: 10.1002/j.1460-2075.1984.tb01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagdasarian M, Timmis KN. Host: vector systems for gene cloning in Pseudomonas. Curr. Top. Microbiol. Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- 47.Katashkina JI, Kuvaeva TM, Andreeva IG, Skorokhodova AY, Biryukova IV, Tokmakova IL, Golubeva LI, Mashko SV. Construction of stably maintained non-mobilizable derivatives of RSF1010 lacking all known elements essential for mobilization. BMC Biotechnol. 2007;7:80. doi: 10.1186/1472-6750-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scherzinger E, Bagdasarian MM, Scholz P, Lurz R, Ruckert B, Bagdasarian M. Replication of the broad host range plasmid RSF1010: requirement for three plasmid-encoded proteins. Proc. Natl Acad. Sci. USA. 1984;81:654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bagdasarian M, Lurz R, Rückert B, Franklin FCH, Bagdasarian MM, Frey J, Timmis KN. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 50.Nishikawa M, Suzuki K, Yoshida K. Structural and functional stability of IncP plasmids during stepwise transmission by trans-kingdom mating: promiscuous conjugation of Escherichia coli and Saccharomyces cerevisiae. Jpn. J. Genet. 1990;65:323–334. doi: 10.1266/jjg.65.323. [DOI] [PubMed] [Google Scholar]
- 51.Waters VL. Conjugation between bacterial and mammalian cells. Nat. Genet. 2001;29:375–376. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- 52.Miller JH. Experiments in Molecular Genetics. NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 53.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1983;1:784–791. [Google Scholar]
- 54.Bussiere D, Bastia D. Termination of DNA replication of bacterial and plasmid chromosomes. Mol. Microbiol. 1999;31:1611–1618. doi: 10.1046/j.1365-2958.1999.01287.x. [DOI] [PubMed] [Google Scholar]
- 55.Salje J. Plasmid segregation: how to survive as an extra piece of DNA. Crit. Rev. Biochem. Mol. Biol. 2010;45:296–317. doi: 10.3109/10409238.2010.494657. [DOI] [PubMed] [Google Scholar]
- 56.Vologodskii A. DNA supercoiling helps to unlink sister duplexes after replication. Bioessays. 2010;32:9–12. doi: 10.1002/bies.200900143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schweder T, Schmidt I, Herrmann H, Neubauer P, Hecker M, Hofmann K. An expression vector system providing plasmid stability and conditional suicide of plasmid-containing cells. Appl. Microbiol. Biotech. 1992;38:91–93. doi: 10.1007/BF00169425. [DOI] [PubMed] [Google Scholar]
- 58.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 59.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tropel D, Van Der Meer JR. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 2004;68:474–500. doi: 10.1128/MMBR.68.3.474-500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diaz E, Prieto MA. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 2000;11:467–475. doi: 10.1016/s0958-1669(00)00126-9. [DOI] [PubMed] [Google Scholar]
- 63.Wieland M, Hartig JS. Artificial riboswitches: rynthetic mRNA-based regulators of gene expression. Chembiochem. 2008;9:1873–1878. doi: 10.1002/cbic.200800154. [DOI] [PubMed] [Google Scholar]
- 64.Gallivan JP. Toward reprogramming bacteria with small molecules and RNA. Curr. Opin. Chem. Biol. 2007;11:612–619. doi: 10.1016/j.cbpa.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Las Heras A, Carreno CA, Martinez-Garcia E, de Lorenzo V. Engineering input/output nodes in prokaryotic regulatory circuits. FEMS Microbiol. Rev. 2010;34:842–865. doi: 10.1111/j.1574-6976.2010.00238.x. [DOI] [PubMed] [Google Scholar]
- 66.Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meighen EA. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 68.Kohlmeier S, Mancuso M, Tecon R, Harms H, van der Meer JR, Wells M. Bioreporters: gfp versus lux revisited and single-cell response. Biosens. Bioelectron. 2007;22:1578–1585. doi: 10.1016/j.bios.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 69.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 70.Miller WG, Lindow SE. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 71.Weaver T, Maurer J, Hayashizaki Y. Sharing genomes: an integrated approach to funding, managing and distributing genomic clone resources. Nat. Rev. Genet. 2004;5:861–866. doi: 10.1038/nrg1474. [DOI] [PubMed] [Google Scholar]
- 72.Mouser PJ, Holmes DE, Perpetua LA, DiDonato R, Postier B, Liu A, Lovley DR. Quantifying expression of Geobacter spp. oxidative stress genes in pure culture and during in situ uranium bioremediation. ISME J. 2009;3:454–465. doi: 10.1038/ismej.2008.126. [DOI] [PubMed] [Google Scholar]
- 73.de Lorenzo V. Recombinant bacteria for environmental release: what went wrong and what we have learnt from it. Clin. Microbiol. Infect. 2009;15(Suppl. 1):63–65. doi: 10.1111/j.1469-0691.2008.02683.x. [DOI] [PubMed] [Google Scholar]
- 74.Ferrer M, Martinez-Abarca F, Golyshin PN. Mining genomes and ‘metagenomes’ for novel catalysts. Curr. Opin. Biotechnol. 2005;16:588–593. doi: 10.1016/j.copbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 75.Wackett LP, Hershberger CD. Biocatalysis and Biodegradation: Microbial Transformation of Organic Compounds. Washington: American Society for Microbiology; 2001. [Google Scholar]
- 76.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]