Abstract
We created SynSysNet, available online at http://bioinformatics.charite.de/synsysnet, to provide a platform that creates a comprehensive 4D network of synaptic interactions. Neuronal synapses are fundamental structures linking nerve cells in the brain and they are responsible for neuronal communication and information processing. These processes are dynamically regulated by a network of proteins. New developments in interaction proteomics and yeast two-hybrid methods allow unbiased detection of interactors. The consolidation of data from different resources and methods is important to understand the relation to human behaviour and disease and to identify new therapeutic approaches. To this end, we established SynSysNet from a set of ∼1000 synapse specific proteins, their structures and small-molecule interactions. For two-thirds of these, 3D structures are provided (from Protein Data Bank and homology modelling). Drug-target interactions for 750 approved drugs and 50 000 compounds, as well as 5000 experimentally validated protein–protein interactions, are included. The resulting interaction network and user-selected parts can be viewed interactively and exported in XGMML. Approximately 200 involved pathways can be explored regarding drug-target interactions. Homology-modelled structures are downloadable in Protein Data Bank format, and drugs are available as MOL-files. Protein–protein interactions and drug-target interactions can be viewed as networks; corresponding PubMed IDs or sources are given.
INTRODUCTION
Synapses are specialized subcellular organelles connecting nerve cells in the central nervous system or nerve cells and muscle cells in the peripheral nervous system. Synapses act by pre-synaptic release of synaptic transmitters and the post-synaptic reception of the signal. Their outstanding significance for learning was explained by Donald Hebb in 1949 with the phrase: ‘cells that wire together, fire together’ (1). This concept of synaptic plasticity means that repetitive correlated firing between neurons leads to improved transmission between those neurons. To date, important details of the underlying molecular mechanisms involving the glutamate receptors and downstream signalling proteins are known (2). More than 100 neurological diseases, such as autism or schizophrenia, are associated with mutations of synaptic proteins (3), which may represent novel therapeutic targets. Proteomic data analysis may allow correlations between mutations in synaptic proteins and monogenic diseases to be found as demonstrated by Bayes et al. (4). Using the international classification of diseases (ICD-10), they found that the terms ‘psychiatry’ and ‘neurology’ (Chapters V and VI) were predominant for synaptic proteins, which includes neurodegenerative diseases like Parkinson’s or Huntington’s, mental retardation and motor disorders, such as dystonia or epilepsies.
A number of resources dedicated to the synapse were developed and are listed in the Society for Neuroscience Information Framework (http://neuinfo.org). More specifically, the SynDB (5), based on a synaptic ontology, created a resource for 14 000 synaptic proteins (3000 in humans). The Genes to Cognition database/SYNSYSdb contains 5000 mammalian genes for synapse proteins and associated information, including mutations, interactions and so forth, aiming at warehousing data on the synaptic proteome (6). Recently, the gene-centred database SynaptomeDB compiled 1900 human synaptic genes (7) that were linked to STRING (8), which provides information about experimental and predicted protein–protein interactions (PPIs).
Here, we present SynSysNet, which is based on an expert-curated list of 1000 human genes that are specific to the synapse. Information on the resulting proteins, their 3D structure, small molecules that bind to them and PPIs was integrated. SynSys stands for ‘Synaptic Systems’ including synaptic gene function in different animal models, and the SynSys consortium is dedicated to systems biology of the synapse (http://synsys.eu; http://synsysdb.genes2cognition.org) using various experimental techniques to elucidate synaptic PPIs (9) and to develop disease-oriented models. Therefore, the integration of a confidence score for experimental results such as the one developed in HIPPIE (10) was important. To bridge the gap between interaction proteomics and diseases, it will be important to consider a therapy-oriented drug classification, which allows a mapping of drug-target relations onto the PPI network of the synapse. To this end, the hierarchical World Health Organization classification assigning Anatomical Therapeutic Chemical codes to drugs (ATC-codes) (11) was implemented in SynSysNet. The complexity and importance of the synaptic system, which involves a large number of specific proteins and has been a target of more drugs than any other human tissue, requires integrative resources combining multiple data and analyses. SynSysNet was developed to aid researchers to integrate current structural and interaction data on synaptic proteins with the aim of understanding the effect of existing and potentially novel drug therapies.
MATERIALS AND METHODS
Manual curation of the list of synaptic genes was a time-consuming step, requiring discussion between different laboratories. Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability (12). The goal was the selection of the proteins that occur specifically in the synapse. This excluded, for example, the products of housekeeping genes, including some prominent proteins of mitochondria, energy production or carbohydrate metabolism, but generated a more focused data set. UniProt mapping of gene names to proteins (13) was used to identify the associated proteins, and splice forms were obtained from Ensembl (14). Drug-target relations were integrated from SuperTarget (15) for all proteins from the list of the SynSys consortium.
SynSysNet integrates PPIs with experimental evidence. In contrast to other resources, predicted interactions are not included in the database. Instead, there are two resources, data from the SynSys consortium and articles on experimental work, where only data with PubMed IDs are displayed. Future releases of the database will integrate new data from consortium partners. Automatic text mining and Web-resources [see (16) for a list] were used to generate a PPI network, which can be evaluated by a normalized quality scoring scheme (10). This quality score is computed using a function that takes into account three components: the number of studies that report a given PPI, the type of technique used for the detection of the PPI and the number of species in which orthologues of the human PPI partners have been experimentally verified to interact. The evaluation of the techniques was done in collaboration with experts in the production of PPI data, and more direct methods were scored more highly (e.g. X-ray crystallography). The function was optimized to score reproducible interactions most highly using leave-one-out re-sampling. In its SynSys implementation, the original HIPPIE scores are rescaled to integers between 0 and 10; values of ≥5 can be considered to indicate high-confidence interactions. For details about the HIPPIE score implementation see the frequently asked questions on the website.
Server, database and system requirements
SynSysNet is a relational MySQL (http://www.mysql.com) database. To handle the chemical information within the database, the MyChem package (http://mychem.sourceforge.net/) is used. MyChem relies for most of its functions on the Open Babel toolbox (http://openbabel.org). The website is built using PHP (http://www.php.net/); web access is enabled through Apache HTTP Server (http://httpd.apache.org/). We recommend a recent version of Mozilla Firefox; alternate browsers Google Chrome, Microsoft Internet Explorer and Apple Safari were tested with some configurations. JavaScript must be enabled, as Java is required to use all features of the site.
3D structures and homology models
To obtain 3D structures for the SynSys proteins and template structures for modelling, a Basic Local Alignment Search against the Protein Data Bank (PDB) (17) was performed. Here, all non-redundant PDB sequences with a sequence identity of 95% were selected. Modeller 9v10 was used for the homology modelling. The generation of each model is based on the PDB structure selected according to identity, alignment length and E-value. The quality of the 3D model was evaluated by the DOPE (18) score, a DOPE-score plot for each model is provided.
Visualization
The network of synaptic interactions, including drugs and proteins, can be displayed interactively through CobWeb (19), an open source Java applet for network exploration. Pathway information is acquired from KEGG (20) through Web service, and drug-target information is shown on top in a pop-up layer. Protein structures can be visually inspected using Jmol (21).
RESULTS AND DISCUSSION
Synaptic genes were manually curated to assure that proteins were, based on PubMed data, correctly annotated as synaptic. General ‘housekeeping’ enzymes (e.g. for energy production or carbohydrate metabolism) were excluded to obtain a focused data set. These criteria led to a list of 1028 synaptic genes. The genes and associated proteins (including synonyms, splice forms and so forth) are available in the download section of the database. Revised/extended versions of the list will be released in upcoming versions of SynSysNet.
A total of 46 000 compounds were associated with the synaptic proteins. The database comprises 69 000 compound-target relations. The corresponding PubMed ID or resource can be retrieved by clicking on the edge between targets in the network-view. For most of the interactions, quantitative binding values are available. A subset of 750 approved drugs (with ATC codes) addresses synaptic targets. Although 30% of all drugs interact with synaptic targets, >60% of all ‘N-drugs’ (ATC code N, nervous system) occur in the network of synaptic proteins. Other prominent ATC classes represented include C (cardiovascular, 40%), S (sensory organs, 40%) and R (respiratory, 28%). Some of the synaptic targets that cause this distribution of drug frequencies in the network can be identified in Figure 1. Well-represented drug classes in SynSysNet are psycholeptics with 90 drugs (ATC code: N05), ophthalmologicals with 72 drugs (S01), psychoanaleptics with 66 drugs (N06), cardiac therapeutics with 48 drugs (C01), analgesics with 40 drugs (N02), drugs for obstructive airway diseases with 32 agents (R03), anaesthetics with 29 drugs (N01), anti-epileptics with 27 drugs (N03) and other nervous system drugs with 24 representatives (N07).
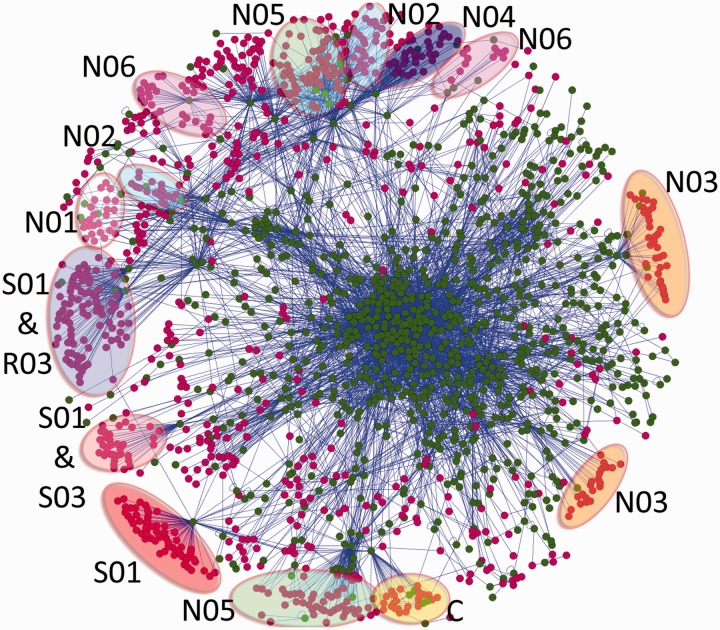
Figure 1.
Drug–PPI network. This compound–PPI network shows the interactions between 1160 compounds and 894 proteins. To create the network, compounds and connected proteins from SynSysNet were chosen. The proteins, the compounds and the interactions were loaded into Cytoscape (22); a graph layout minimizing the total path length (spring embedded, edge-weighted layout) was used. A total of 6116 interactions (edges), composed of 4318 PPIs and 1798 compound–protein interactions, are shown. Compounds are represented by ruby circles and proteins by green circles, and the edges between them are blue. Some compounds have the same target; therefore, they are clustered into groups. The clusters were analysed, and they were found to contain many compounds belonging to the same ATC code (highlighted). Most of the clusters comprise drugs of the nervous system (N) ATC group. The N01 cluster, which represents anaesthetics, is highlighted white, N02 (analgesics) light-blue, N03 (anti-epileptics) orange, N04 (anti-Parkinson’s drugs) dark-blue, N05 (psycholeptics) green and N06 (psychoanaleptics) pink. Besides the nervous system ATC codes, drugs from other ATC groups occur, sensory organs (S), respiratory system (R) and cardiovascular system (C). There are S01 codes (ophthalmologicals) highlighted in red, S01 and S03 (ophthalmological and otological preparations) combined in red violet, S01 and R03 (drugs for obstructive airway diseases) combined in purple and the cardiovascular system (C) in yellow.
To date, 5000 PPIs, referenced by 8000 literature references, can be explored; the quality score is used to draw the thickness of the network edges in the interactive network visualization. PPIs of the synapse generate a complex dynamic network. Comprehension of this network will be important for a deeper functional understanding of the synapses and their related diseases, the so-called synaptopathies (3).
Compound PPIs are visualized in Figure 1. SynSysNet provides interactive graphical Web access to these data, allowing users to extend or reduce a network that can then be exported in an XML format [XGMML, standard in Cytoscape (22)]. Although, as it could be expected, the drugs of the nervous system dominate this network, other main classes of the ATC tree are represented. Visual examination of the unbiased network graph suggests that most of the approved drugs and their targets occur in the outer shell of the graph. As proteins with many interactions cluster in the centre of the graph, this could have been caused by the fact that drugs target preferentially synapse proteins with a small number of interactions. However, we could not confirm a significant difference in the degree distribution of protein interactions of the proteins targeted by drugs versus those not targeted (data not shown). This is in contrast to the observation that cancer drugs target high degree proteins (23); this could be taken as an indication that drug–protein networks related to different tissues or diseases may have different topological properties and require different approaches.
An interesting feature evidenced by the drug–protein graph is the existence of clusters of drugs. The reason is that the majority of the drugs belonging to the same ATC class share at least one target. Thereby those drugs accumulate around this target and are located close to each other. For instance, the N05 class contains psycholeptics, which have an anti-psychotic, anxiolytic and sedative effect, and generally target central nervous system function. In Figure 1, the N05 class is divided into two groups and each addresses a different 5-hydroxytryptamine (serotonine) receptor. The lower group seems to be more concentrated on one target the gamma-aminobutyric acid (GABA) receptor, whereas the upper group in the figure shares many interaction partners with other ATC classes (N02, N04, N06). One neighbour of the upper N05 group is N04, which acts against Parkinson’s disease through an activation of the dopaminergic system. It is plausible that these share targets because it is known that psycholeptics inhibit the dopaminergic system and often lead to undesirable side effects, such as Parkinson’s-like symptoms. Accordingly, it can be assumed that the lower N05 group will cause less Parkinson’s-like adverse effects. Another example that explains drug side effects on a molecular level is the lower N03 cluster (anti-epileptics), which addresses a primary target (sodium channel). This explains the therapeutic effect and the side effect hypertension because the drug targeted sodium channel is also expressed in the cardiac muscle.
As aforementioned, other ATC codes appear in this synaptic network in addition to the N ATC group for drugs, which have an effect in the nervous system. These other drugs belong to the S (sensory organs), R (respiratory system) and C (cardiovascular system) ATC groups. This presence can be explained by their interacting targets, which are not only expressed in the central nervous system but also in other tissues. For example, the S01 and R03 groups share the M1- and M2-type muscarinic acetylcholine receptor targets. These receptors have physiological functions in different tissues; therefore, they occur in the central nervous system as well as in lung and sensory organs. The tissue distribution of these receptors is important for the S01 and R03 groups, which contain ophthalmologicals (S01) and drugs for obstructive airway diseases (R03). Therefore, they are only used topically and do not affect the muscarinic acetylcholine receptor in the brain.
Another example is the ATC group C, which contains drugs against cardiovascular diseases. Some of those drugs inhibit the Na(+)/K(+)-adenosine triphosphatase, which plays a major role in the physiological function of every cell and is highly important for neuronal function. As this transporter is broadly expressed, these drugs have a narrow therapeutic window. For example, one of these C-group drugs, digitoxin, causes abnormal colour vision as a side effect, possibly by affecting synapses in the central nervous system.
SynSysNet was developed as a user-friendly platform, and Figure 2 shows the use of some features of the website exemplified by exploration of the Parkinson’s drug–PPI network. The navigation bar in the upper area offers downloads, answers to frequently asked questions, links and so forth.
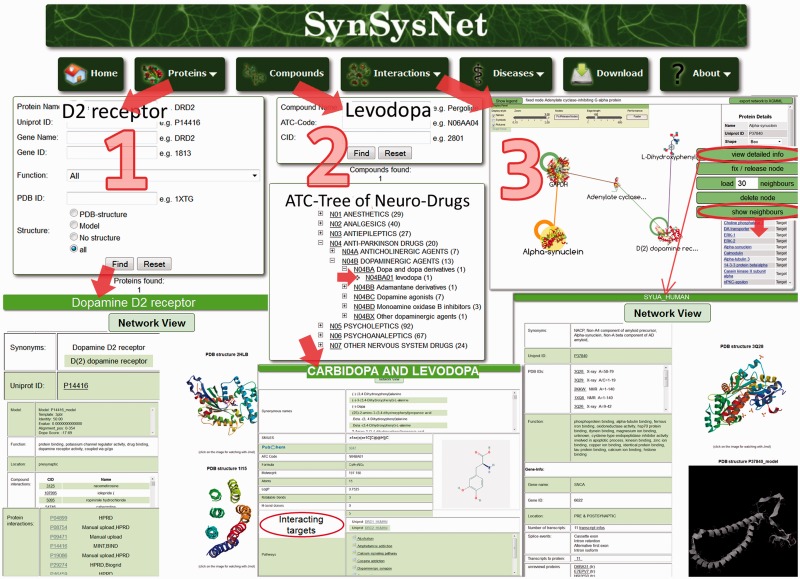
Figure 2.
A SynSysNet example query. Step 1 shows the search for a protein, in this example the D2 dopamine receptor. By clicking on the ‘Find’ button, the result list will open on the next page with detailed information like synonyms, IDs, function, gene information, pathways and PDB structures/3D models. The button ‘Network view’ leads to an interactive network visualized by CobWeb, which shows the interacting proteins and drugs. Step 2 illustrates the search for a compound either by name, for example, l-dopa, or by the ATC code, for example, N04BA01. The results will be presented on the next page, again with detailed information about the compound, particularly the interacting targets, in this case D(1A) and D(2) dopamine receptor. Finally, in Step 3, an interactive network from the disease, for example, ‘Parkinson’s disease’, is shown. For every protein and compound, detailed information is provided (see the button “view detailed info”), including complete ‘neighbourhood’ (drugs and targets).
There are three ways to explore a drug–PPI network:
The ‘Proteins’ button, illustrated in Step 1, enables the user to type in a protein name or common IDs (e.g. the dopamine D2 receptor, which is a major target of the medication for Parkinson’s disease) or any ID. It is also possible to search by the function of a protein. Clicking on ‘Find’ leads to a list of detailed information on the protein, including localization, 3D structures. Clicking on the button ‘Network view’ on the top of the page leads to the compound–PPI network, as shown in Step 3.
The user is able to search for compounds as well as ATC codes (e.g. l-dopa or N04BA01) by clicking on the ‘Compounds’ button. Information about the drug l-dopa is shown in Step 2. It is possible to get a ‘network view’ or to get information on ‘interacting targets’ or ‘pathways’ by clicking on these links. The occurrence of synaptic proteins in ∼200 KEGG-pathways and in 30 synapse specific pathways can be explored, and the binding compounds can be displayed (for details see Figure 3).
‘Interactions’ are illustrated in Step 3, where the user is able to directly search for a specific network by a number of drugs/targets or predefined disease networks. The example shows the network of Parkinson’s disease. The dopamine receptor is shown with 30 neighbours (default: 15 targets and 15 drugs); all neighbours can be listed by clicking on ‘show neighbours’, and a distinct number can be added by clicking ‘load x neighbours’ on the right side of the network. Detailed information on specific targets (e.g. α-synuclein) can be viewed in a separate tab. The dopamine receptors are surrounded by a number of drugs in the right upper corner of the network. Most of them act on both D(1) and D(2) receptors. The relation to other mechanisms of action, like inhibition of the glutamate- (NMDA), monoamine-oxidase- (MAO-A/B) and muscarinergic receptors, can be explored in this graph. Corresponding drugs (amantadine, rasagiline and biperiden) and other compounds are displayed. α-Synuclein is a soluble protein that regulates dopamine secretion and generates membrane channels. Mutations are responsible for different versions of Parkinson’s disease and so-called Lewy-bodies, which are synuclein-inclusions. α-Synuclein automatically appears in the SynSys drug–PPI network of Parkinson’s disease. α-Synuclein is the promising target of a novel vaccination therapy against Parkinson’s disease that has reached the phase of clinical trials (24). Other promising targets like dopamine transporter or tyrosine hydroxylase and associated compounds can be identified in the SynSys drug–PPI network.
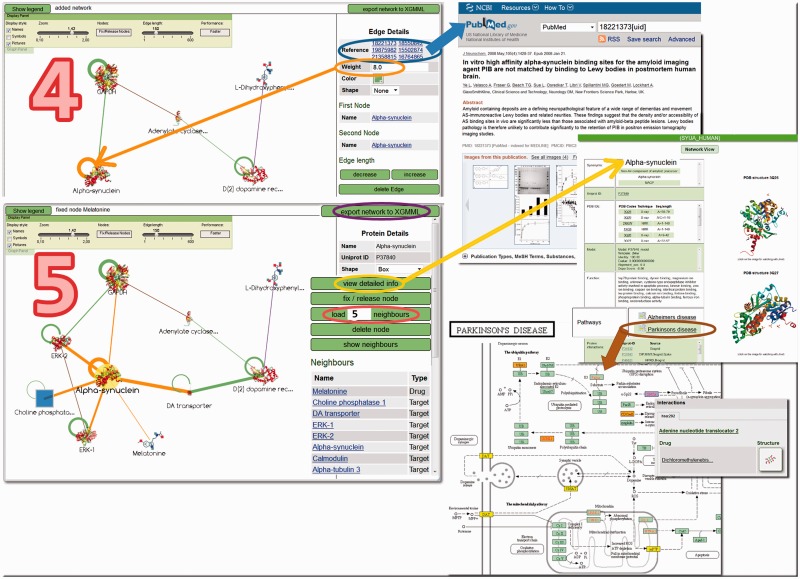
Figure 3.
Network exploration options. Step 4 shows the example of a subnetwork for α-synuclein. On the right side, references with links to PubMed are displayed. The circles correspond to self-interactions. The weight of 8.0 and the circle in the graph correspond to the experimental confidence score of synuclein–synuclein interactions (see ‘Materials and Methods’ section). In Step 5, additional interactors of α-synuclein were loaded. The network can be exported in its current form in XGMML format through the button in the upper right corner. Detailed information on the compounds can be displayed in a new tab. At the bottom of the details page, pathways for different diseases are listed and can be viewed as shown here for Parkinson’s disease (enriched KEGG pathway). Yellow boxes indicate KEGG targets with drug info in SynSysNet. Mouse-over results in a tool-tip (pop-up) that contains detailed drug-target information.
Some network exploration options are illustrated in Figure 3, with the examples of α-Synuclein and the dopamine receptor. In Step 4, clicking on the edges of the interaction graph gives PubMed IDs (clickable) or resource and experimental confidence scores (weight). For the target under examination, more neighbours can be loaded into the visualization (individually from a list or a defined number). The ‘view detailed info’ option leads to more information, including KEGG pathways, which complement the network view because here, the targets are grouped according to functional or pathological units.
Recently, it was shown that PPI networks could be exploited to find novel targets and to judge their druggability (25,26). The mapping of drug-target relations onto the network of synaptic PPIs will enable new questions to be asked, for instance: what is the reason for an adverse effect of a drug in the (central) nervous system? The value of high quality PPIs for such systems-level understanding of disease and therapy cannot be overestimated (27). Data like localization, dynamics and quantitative aspects like stoichiometry and binding affinities will extend existing PPIs (28). The yet preliminary data on pre-/post-synaptic and vesicular localization will be scrutinized during the project, data on PPIs will grow, new disease models will be developed and known 3D structures of synapse proteins will be used to generate larger protein complexes according to experimental data.
Synaptic proteins and synaptic protein interactions are crucially contributing to synaptic function and plasticity in health and disease. Given the socio-economic impact of brain disease and the substantial role of synaptopathies in these, we expect that specifically targeting certain synaptic proteins, or synaptic protein interactions, will become highly important in the years to come.
The newly established compound–PPI network will aid the understanding of complex interactions between transmitters, drugs and synaptic proteins, which could explain various pathological conditions as well as the adverse effects of drugs. SynSysNet will gain more predictive value as more data on PPIs will become known. Notably, large-scale efforts to map the synaptic interactome are ongoing and will be used to continuously update SynSysNet.
AVAILABILITY
SynSysNet is available through the website without registration: http://bioinformatics.charite.de/synsys. It can be used under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 License.
FUNDING
European Union Seventh Framework Programme SYNSYS (Synaptic Systems: dissecting brain function in health and disease), HEALTH-2009-2.1.2-1, under grant agreement no [242167]; Deutsche Forschungsgemeinschaft [GRK1772, GRK1360]. Funding for open access charge: European Union, FP7, SynSys.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the colleagues of the SynSys consortium for the contribution of experimental data on PPIs.
REFERENCES
- 1.Whitcomb DJ, Regan P, Cho K. The synapse and brain function. Semin. Cell Dev. Biol. 2011;22:488–491. doi: 10.1016/j.semcdb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Yuzaki M. New (but old) molecules regulating synapse integrity and plasticity: Cbln1 and the delta2 glutamate receptor. Neuroscience. 2009;162:633–643. doi: 10.1016/j.neuroscience.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Grant SG. Synaptopathies: diseases of the synaptome. Curr. Opin. Neurobiol. 2012;22:522–529. doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Bayes A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, Grant SG. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Zhang Y, Zheng H, Zhang C, Xiong W, Olyarchuk JG, Walker M, Xu W, Zhao M, Zhao S, et al. SynDB: a Synapse protein DataBase based on synapse ontology. Nucleic Acids Res. 2007;35:D737–D741. doi: 10.1093/nar/gkl876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the genes to cognition database. Nucleic Acids Res. 2009;37:D846–D851. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirooznia M, Wang T, Avramopoulos D, Valle D, Thomas G, Huganir RL, Goes FS, Potash JB, Zandi PP. SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics. 2012;28:897–899. doi: 10.1093/bioinformatics/bts040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li KW, Klemmer P, Smit AB. Interaction proteomics of synapse protein complexes. Anal. Bioanal. Chem. 2010;397:3195–3202. doi: 10.1007/s00216-010-3658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaefer MH, Fontaine JF, Vinayagam A, Porras P, Wanker EE, Andrade-Navarro MA. HIPPIE: integrating protein interaction networks with experiment based quality scores. PLoS One. 2012;7:e31826. doi: 10.1371/journal.pone.0031826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO Expert Committee. The selection and use of essential medicines. World Health Organ. Tech. Rep. Ser. 2009: 1–242. [PubMed] [Google Scholar]
- 12.Ruano D, Abecasis GR, Glaser B, Lips ES, Cornelisse NL, de Jong APH, Evans DM, Davey SG, Timpson NJ, Smit AB, et al. Interaction proteomics of synapse protein complexes. Am. J. Hum. Genet. 2010;86:113–125. doi: 10.1016/j.ajhg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecker N, Ahmed J, von Eichborn J, Dunkel M, Macha K, Eckert A, Gilson MK, Bourne PE, Preissner R. SuperTarget goes quantitative: update on drug-target interactions. Nucleic Acids Res. 2012;40:D1113–D1117. doi: 10.1093/nar/gkr912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathur A, Shankaracharya, Vidyarthi AS. SWIFT MODELLER v2.0: a platform-independent GUI for homology modeling. J. Mol. Model. 2012;18:3021–3023. doi: 10.1007/s00894-011-1319-6. [DOI] [PubMed] [Google Scholar]
- 19.von Eichborn J, Bourne PE, Preissner R. Cobweb: a Java applet for network exploration and visualisation. Bioinformatics. 2011;27:1725–1726. doi: 10.1093/bioinformatics/btr195. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson RM. Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. 2010;43:1250–1260. [Google Scholar]
- 22.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hase T, Tanaka H, Suzuki Y, Nakagawa S, Kitano H. Structure of protein interaction networks and their implications on drug design. PLoS Comput. Biol. 2009;5:e1000550. doi: 10.1371/journal.pcbi.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneeberger A, Mandler M, Mattner F, Schmidt W. Vaccination for Parkinson's disease. Parkinsonism Relat. Disord. 2012;18(Suppl. 1):S11–S13. doi: 10.1016/S1353-8020(11)70006-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, Su S, Bhatnagar RK, Hassett DJ, Lu LJ. Prediction and analysis of the protein interactome in Pseudomonas aeruginosa to enable network-based drug target selection. PLoS One. 2012;7:e41202. doi: 10.1371/journal.pone.0041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Las Rivas J, Prieto C. Protein interactions: mapping interactome networks to support drug target discovery and selection. Methods Mol. Biol. 2012;910:279–296. doi: 10.1007/978-1-61779-965-5_12. [DOI] [PubMed] [Google Scholar]
- 27.Stelzl U, Wanker EE. The value of high quality protein-protein interaction networks for systems biology. Curr. Opin. Chem. Biol. 2006;10:551–558. doi: 10.1016/j.cbpa.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Sorokina O, Sorokin A, Armstrong JD. Towards a quantitative model of the post-synaptic proteome. Mol. Biosys. 2011;7:2813–2823. doi: 10.1039/c1mb05152k. [DOI] [PubMed] [Google Scholar]