Abstract
InnateDB (http://www.innatedb.com) is an integrated analysis platform that has been specifically designed to facilitate systems-level analyses of mammalian innate immunity networks, pathways and genes. In this article, we provide details of recent updates and improvements to the database. InnateDB now contains >196 000 human, mouse and bovine experimentally validated molecular interactions and 3000 pathway annotations of relevance to all mammalian cellular systems (i.e. not just immune relevant pathways and interactions). In addition, the InnateDB team has, to date, manually curated in excess of 18 000 molecular interactions of relevance to innate immunity, providing unprecedented insight into innate immunity networks, pathways and their component molecules. More recently, InnateDB has also initiated the curation of allergy- and asthma-related interactions. Furthermore, we report a range of improvements to our integrated bioinformatics solutions including web service access to InnateDB interaction data using Proteomics Standards Initiative Common Query Interface, enhanced Gene Ontology analysis for innate immunity, and the availability of new network visualizations tools. Finally, the recent integration of bovine data makes InnateDB the first integrated network analysis platform for this agriculturally important model organism.
INTRODUCTION
The innate immune response is a critical branch of immunity, which not only provides a first line of defence against pathogens, but also regulates and shapes subsequent adaptive responses. Innate immunity, however, can also do great harm by driving inappropriate inflammatory cascades. Therefore complex molecular networks are required to regulate innate immunity and maintain appropriate and specific responses to different pathogens, while limiting potential harm from dysregulated inflammation (1–5). The intricate interplay of a multitude of regulatory layers that initiate and coordinate the innate immune response has led to an ever-increasing interest in applying systems-oriented approaches to better understand innate immunity and its modulators (6).
InnateDB (publicly accessible at http://www.innatedb.com and mirrored at http://innatedb.teagasc.ie) is a knowledge base and analysis platform that was specifically designed to provide a system-oriented yet user-friendly tool for integrative analyses of the mammalian innate immune response (7).
INNATEDB CURATION
A key component of the InnateDB project is the contextual manual curation of innate immunity interactions, pathways and their component molecules. The curation process has previously been described in detail (8). InnateDB was first publicly released in 2008 (7). At that time, ∼3500 molecular interactions had been curated. By 2010, the database contained 11 786 InnateDB-curated molecular interactions from the review of >3000 published articles. As of September 2012, our curation team has reviewed >4300 publications, and >18 000 interactions of relevance to innate immunity have been annotated (Figure 1; for detailed statistics see http://www.innatedb.com/statistics). More recently, as part of the AllerGen Networks of Centres of Excellence (NCE) (http://www.allergen-nce.com), InnateDB curators have also begun to annotate interactions and pathways of relevance to allergy and asthma. All interactions in InnateDB are provided with detailed contextual information according to the minimum information required for reporting a molecular interaction experiment (MIMIx) standards (9), including the evidence supporting each interaction, the tissue or cell type the interaction was reported in, the type of interaction and the method of detection. New interactions are added to the database weekly, providing up-to-date annotation on the innate immunity interactome. This resource can be mined to identify new relationships between innate immunity and other processes, to identify potential novel regulators of innate immunity and to interpret a user’s own data (e.g. gene expression data) from a network biology perspective.
Figure 1.
The InnateDB curated interactome in July 2012. Red edges represent interactions that have been added in 2011 and 2012.
Building a comprehensive list of innate immunity genes
Aside from annotating molecular interactions, InnateDB now also annotates which genes have a published role in innate immunity, providing a brief summary of that role and links to the relevant publications. This data set, available at http://www.innatedb.com/curatedGenes, presently contains >1500 genes (957 human, 527 mouse and 46 bovine) and is the most comprehensive list of genes involved in innate immunity that is available. This list was recently used by a group of researchers to show that human proteins, which are targeted by viruses, are highly enriched for having a role in innate immunity (10).
Contribution to the International Molecular Exchange Consortium
In 2010, InnateDB became a member of the International Molecular Exchange Consortium (IMEx) (11). This organization is dedicated to developing rules for describing molecular interaction data, actively curating these interactions from the scientific literature and making them available through a common website.
Within IMEx, InnateDB has committed to curating every issue of Nature Immunology from September 2010 onwards using IMEx curation standards (12). Because IMEx curation requires more annotation detail than the MIMIx level (9) currently supported by InnateDB’s submission system, InnateDB curators are submitting these IMEx interactions through the IntAct interaction database (13). On submission, each IMEx interaction is thoroughly reviewed by an IntAct curator before it is accepted and released. In addition to submitting to IntAct, all InnateDB acceptable interactions (i.e. interactions of relevance to innate immunity) from Nature Immunology are also deposited into InnateDB.
Integrating data from external resources
To supplement our manual curation efforts and to provide a snapshot of the entire interactome beyond known innate immunity interactions, InnateDB imports data from a wide range of genome, interaction and pathway databases (http://www.innatedb.com/resources). Currently, InnateDB contains 178 000+ imported experimentally validated interactions, 3000+ pathways and 300 000+ interactions based on Ortholuge (14) orthology predictions (interologs) in addition to the 18 000+ InnateDB manually curated interactions.
INTEGRATION OF BOVINE DATA—ORTHOLOGY-BASED PATHWAY & NETWORK RECONSTRUCTION
In February 2012, a new version of InnateDB was released that included the incorporation of bovine gene, pathway and molecular interaction annotation in addition to the existing data for human and mouse. This new version of the platform now also facilitates a systems biology approach to the investigation of the bovine innate immune response and is poised to deepen our understanding of important bovine infectious diseases associated with significant economic losses (e.g. bovine tuberculosis and mastitis), as well as enabling cross-species comparisons of innate immunity.
As bovine experimentally validated interactions and pathways are virtually non-existent, InnateDB uses an orthology-based approach to predict bovine pathways and interactions primarily from human data. One should be aware that this approach results in a humanized and frequently incomplete representation of the bovine interactome, but in the absence of widespread experimental data it provides at least a network biology framework to build on and to generate hypotheses that can be subsequently experimentally validated. InnateDB experimentally validated and predicted interactions are clearly labelled. As of September 2012, InnateDB contains >70 000 bovine interologs (interactions based on orthology) involving 10 717 bovine genes. In each case, one can link back to the orthologous human interaction to review evidence for the interaction.
The latest release of InnateDB also uses orthology predictions to transfer human and mouse pathway annotations to bovine genes in real time. Currently, pathway annotations can be assigned to 7032 bovine genes by orthology to human genes. Notably, although only ∼70% of all human genes (14 316 genes) have a predicted bovine ortholog, and a significantly higher proportion (85%) of human genes with pathway annotations have a bovine ortholog. This higher prevalence of conserved genes among pathway-annotated genes indicates that many of the associated processes may be well preserved.
To further examine the appropriateness of the orthology-based annotation transfer on a per-pathway basis, we determined the ‘conservation rate’ (cons) of each pathway, defined as the ratio of pathway participants in the source organism (human/mouse) that have a putative counterpart in the target organism (cow) to the total number of participants in the source organism. As of September 2012, InnateDB contains 1536 human pathways with five or more pathway participants, 80% (1257 pathways) of these have a conservation rate of 0.8 or better. The corresponding number for a conservation rate of ≥0.7 is 93% (1442 pathways). The high prevalence of strongly conserved pathways seems to largely justify an orthology-based approach for inferring bovine pathways. Supplementary Table S1 lists the remaining 107 pathways with a relatively low conservation rate (cons <0.7). Notably, the list of pathways for which an orthology-based reconstruction is challenging includes >30 immunologically important pathways. In some cases, the low conservation rate can be attributed to real divergence of the underlying processes. The bovine Type I Interferon family, for example, has been shown to have undergone widespread expansion, including the divergence of a new Type I interferon (IFN) family (IFNX) in the cow from IFN alpha (15). In other cases, the conservation rate might further increase with future improvements to the quality of the bovine draft genome.
In addition to orthology-based annotation transfer, the tissue expression profile of a gene can provide some insight into its potential function (16). Through collaboration with colleagues at the United States Department of Agriculture, InnateDB now integrates bovine tissue expression data for >13 000 genes. This data was sourced from the Bovine Gene Atlas (17), which has profiled gene expression across 87 different bovine tissues using a next generation sequencing approach. A graphical tissue expression profile is available on the Gene Card page of bovine genes.
INNATEDB DATA ANALYSIS AND VISUALIZATION
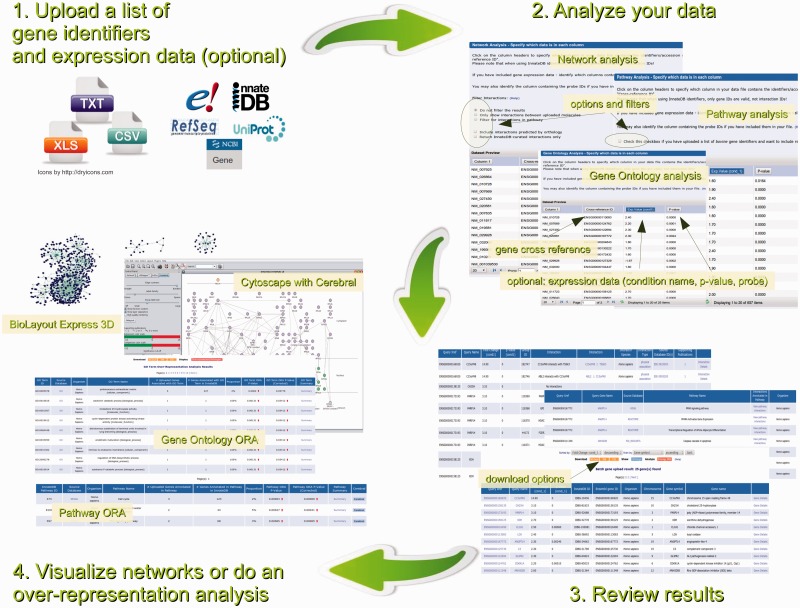
InnateDB can serve as a knowledge base where users can search for particular genes or proteins of interest and their associated interactions and pathways, using a variety of search fields. Alternatively, InnateDB can be queried in a more high-throughput fashion, where users can upload a list of genes/proteins and associated quantitative data from up to 10 different conditions (e.g. gene expression data) and carry out more complex searches and analyses (Figure 2). After uploading a list of gene IDs (human, mouse and bovine Ensembl, RefSeq, Entrez Gene, UniProt and InnateDB IDs are all accepted), users can quickly find which pathways, Gene Ontologies (including enhanced innate immunity gene annotation) or transcription factor binding sites are statistically over-represented in their dataset. Users can also use InnateDB to build, visualize and analyse molecular interaction networks consisting of their uploaded genes and their encoded products. One can, for example, construct a network of how differentially expressed genes interact with one another. Quantitative data uploaded by the user is automatically overlaid on these networks. Recent improvements to InnateDB include the option to incorporate interactions based on orthology in the construction of molecular interaction networks and the option to restrict the networks to contain only InnateDB manually curated interactions. Further enhancements to the web-interface include more intuitive page layouts, faster searches and analyses, and a variety of other changes (see http://www.innatedb.com/news).
Figure 2.
Data analysis workflow in InnateDB.
Network visualization tools
All interactions in InnateDB may be downloaded in several standardized formats including text-based formats (tab, csv, xls), the simple interaction format (sif) and the PSI-MI XML 2.5 and MITAB formats (18). Additionally, interaction networks may also be visualized in our Cerebral program (19), a Java plugin for the Cytoscape network visualization software (20,21), which uses subcellular localization information to orientate interaction networks in a more biologically intuitive pathway-like layout. Networks can also be visualized in other third-party software including the CyOog plugin (22), which uses Power Graph analysis to reduce network complexity by explicitly representing re-occurring network motifs. Recently, we have also integrated BioLayout Express 3D 2.2 (23), an application designed for the visualization, clustering and analysis of large networks in 2D and 3D space.
Proteomics Standards Initiative Common Query Interface implementation
Interaction data in InnateDB can now also be queried using web services implementing The Proteomics Standards Initiative Common Query Interface (PSICQUIC) (24). PSICQUIC is an effort from the Human Proteome Organization Proteomics Standards Initiative (http://www.hupo.org/research/psi/) to standardize programmatic access to molecular interaction databases based on the PSI standard formats (PSI-MI XML and MITAB) (18). It defines standard web services and also a query syntax for powerful and flexible searches.
All data sources implementing PSICQUIC can be queried in the exact same way, i.e. the same query can be used to retrieve the relevant data from many different interaction data sources. Independently published observations of an experimental system, curated by independent databases, are then integrated in response to a single user query (see http://www.ebi.ac.uk/intact/imex). PSICQUIC web services are RESTful (REpresentational State Transfer) but can also be accessed through SOAP (Simple Object Access Protocol). A list of available services for InnateDB can be found at http://imex.innatedb.com/psicquic-ws/webservices. InnateDB updates the data files for the PSICQUIC web services weekly and additionally provides them for download in a compressed format at http://www.innatedb.com/downloads.
ONGOING DEVELOPMENTS
InnateDB will maintain its curation efforts to annotate interactions and genes of relevance to innate immunity, with weekly updated annotation, thus continuing to provide a comprehensive platform for systems and network biology analyses of innate immune-associated responses. Continued incorporation of data from external resources, encompassing the wider human, mouse and bovine interactomes, will also continue to facilitate analyses beyond innate immunity by a wide range of researchers. Additionally, InnateDB intends to expand beyond the curation of innate immunity relevant networks, incorporating more adaptive immunity information. We are currently developing a first version of an Allergy and Asthma Portal that will further integrate data on allergy and associated immune interactions from both the literature and researchers from AllerGen. This portal will be built on InnateDB and will provide an analysis platform for more sophisticated network biology-based investigations of allergy and asthma responses. These interactions will be identifiable from innate immunity interactions, so that users can continue to have focused analyses on the innate immunity interactome.
Further future developments will include improvements to InnateDB pathway analysis tools. The over-representation–based methods for pathway analysis that are currently available through InnateDB’s data analysis interface are widely established and considered a ‘gold standard’; yet, they neglect the fact that many components are shared between seemingly unrelated pathways. To address this issue, we have used the InnateDB collection of pathway annotations as a basis to identify pairs of genes that co-occur only in a single pathway and developed a novel pathway analysis method [signature over-representation analysis (SIGORA)] that focuses on the over-representation of such gene pairs in a list of genes of interest (Foroushani et al, submitted). SIGORA is currently implemented as an R package [available from the Comprehensive R Archive Network (CRAN) at http://cran.r-project.org/web/packages/sigora/index.html] and will be integrated into the future releases of InnateDB.
Finally, together with the PSICQUIC development team and other IMEx members, we are working on an improved reference implementation of PSICQUIC. We are also preparing to export our data in MITAB 2.7 format.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
This work was supported by Genome BC through the Pathogenomics of Innate Immunity (PI2) project and by the Foundation for the National Institutes of Health and the Canadian Institutes of Health Research under the Grand Challenges in Global Health Research Initiative [Grand Challenges ID: 419]. Further funding was also provided by AllerGen grants 12ASI1 and 12B&B2. D.J.L. was funded in part during this project by a postdoctoral trainee award from the Michael Smith Foundation for Health Research (MSFHR). F.S.L.B. is a MSFHR Senior Scholar and R.E.W.H. holds a Canada Research Chair (CRC). Funding to enable bovine systems biology in InnateDB is provided by Teagasc [RMIS6018] and the Teagasc Walsh Fellowship scheme. IMEx is funded by the European Commission under the PSIMEx project [contract number FP7-HEALTH-2007-223411]. Funding for open access charge: Teagasc [RMIS6018].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Matthew Whiteside for providing orthology predictions in InnateDB. The authors would also like to acknowledge Tad Sonstegard, Lakshmi Matukumalli and other members of the Bovine Gene Atlas project for providing the bovine tissue expression data. We would also like to thank all the previous members of the InnateDB development and curation teams (http://www.innatedb.com/about). Thanks also to the various interaction, pathway and annotation databases that have been integrated into InnateDB for freely providing their data to the public. Grateful thanks also go to the many researchers who have taken the time to respond to our queries regarding curation of their publications.
REFERENCES
- 1.Delano MJ, Thayer T, Gabrilovich S, Kelly-Scumpia KM, Winfield RD, Scumpia PO, Cuenca AG, Warner E, Wallet SM, Wallet MA, et al. Sepsis induces early alterations in innate immunity that impact mortality to secondary infection. J. Immunol. 2011;186:195–202. doi: 10.4049/jimmunol.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi FD, Ljunggren HG, Sarvetnick N. Innate immunity and autoimmunity: from self-protection to self-destruction. Trends Immunol. 2001;22:97–101. doi: 10.1016/s1471-4906(00)01821-4. [DOI] [PubMed] [Google Scholar]
- 5.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardy JL, Lynn DJ, Brinkman FS, Hancock RE. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol. 2009;30:249–262. doi: 10.1016/j.it.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynn DJ, Chan C, Naseer M, Yau M, Lo R, Sribnaia A, Ring G, Que J, Wee K, Winsor GL, et al. Curating the innate immunity interactome. BMC Syst. Biol. 2010;4:117. doi: 10.1186/1752-0509-4-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stumpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 10.Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 11.Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FS, Cesareni G, et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orchard S, Kerrien S, Jones P, Ceol A, Chatr-Aryamontri A, Salwinski L, Nerothin J, Hermjakob H. Submit your interaction data the IMEx way: a step by step guide to trouble-free deposition. Proteomics. 2007;7(Suppl.1):28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- 13.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulton DL, Li YY, Laird MR, Horsman BG, Roche FM, Brinkman FS. Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics. 2006;7:270. doi: 10.1186/1471-2105-7-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker AM, Roberts RM. Characterization of the bovine type I IFN locus: rearrangements, expansions, and novel subfamilies. BMC Genomics. 2009;10:187. doi: 10.1186/1471-2164-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dezso Z, Nikolsky Y, Sviridov E, Shi W, Serebriyskaya T, Dosymbekov D, Bugrim A, Rakhmatulin E, Brennan RJ, Guryanov A, et al. A comprehensive functional analysis of tissue specificity of human gene expression. BMC Biol. 2008;6:49. doi: 10.1186/1741-7007-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harhay GP, Smith TP, Alexander LJ, Haudenschild CD, Keele JW, Matukumalli LK, Schroeder SG, Van Tassell CP, Gresham CR, Bridges SM, et al. An atlas of bovine gene expression reveals novel distinctive tissue characteristics and evidence for improving genome annotation. Genome Biol. 2010;11:R102. doi: 10.1186/gb-2010-11-10-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon—level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barsky A, Gardy JL, Hancock RE, Munzner T. Cerebral: a cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics. 2007;23:1040–1042. doi: 10.1093/bioinformatics/btm057. [DOI] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royer L, Reimann M, Andreopoulos B, Schroeder M. Unraveling protein networks with power graph analysis. PLoS Comput. Biol. 2008;4:e1000108. doi: 10.1371/journal.pcbi.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout express(3D) Nat. Protoc. 2009;4:1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 24.Aranda B, Blankenburg H, Kerrien S, Brinkman FS, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]