Abstract
The ESTHER database, which is freely available via a web server (http://bioweb.ensam.inra.fr/esther) and is widely used, is dedicated to proteins with an α/β-hydrolase fold, and it currently contains >30 000 manually curated proteins. Herein, we report those substantial changes towards improvement that we have made to improve ESTHER during the past 8 years since our 2004 update. In particular, we generated 87 new families and increased the coverage of the UniProt Knowledgebase (UniProtKB). We also renewed the ESTHER website and added new visualization tools, such as the Overall Table and the Family Tree. We also address two topics of particular interest to the ESTHER users. First, we explain how the different enzyme classifications (bacterial lipases, peptidases, carboxylesterases) used by different communities of users are combined in ESTHER. Second, we discuss how variations of core architecture or in predicted active site residues result in a more precise clustering of families, and whether this strategy provides trustable hints to identify enzyme-like proteins with no catalytic activity.
INTRODUCTION
The α/β-hydrolase fold family of enzymes described in 1992 (1) is rapidly becoming one of the largest groups of structurally related proteins with diverse catalytic and non-catalytic functions. The fold is composed of a β-sheet of eight strands with the second strand antiparallel to the others and ordered as 12435678. The prototype of enzymes in the fold has a catalytic triad composed of a nucleophilic residue located at the top of a γ-turn between the fifth β-strand and the following α–helix (the nucleophile elbow), an acidic (Glu or Asp) residue and a His. Since the initial description of this fold, few reviews have dealt with the increasing number of functions performed by members of the family (2–5). The ESTHER database, dedicated to proteins with an α/β-hydrolase fold, was created in 1995 (6), and since then, it has been upgraded regularly (7–11), for example, with incorporation of new tools serving different fields of research, such as analysis of mutations in humans associated with diseases (12) or mutations in pests developing resistance to insecticides (13).
Catalytic members (enzymes) in this superfamily include hydrolases (acetylcholinesterase, carboxylesterase, dienelactone hydrolase, lipase, cutinase, thioesterase, serine carboxypeptidase, proline iminopeptidase, proline oligopeptidase, epoxide hydrolase) along with enzymes that require activation of HCN, H2O2 or O2 instead of H2O for the reaction mechanism (haloalkane dehalogenase, haloperoxidase, hydroxynitrile lyase) (14). Known non-catalytic members include the neuroligins, glutactin, neurotactin, the C-terminal domain of thyroglobulin, yolk proteins, the CCG1-interacting-factor-B and dipeptidylaminopeptidase VI. Some proteins are suspected to display more than one function and could be true moonlighting proteins (15). As the vast majority of members have not yet been characterized experimentally, many more new functions could emerge in the least known subfamilies (e.g. the domains of unknown function, or DUF) in the future. The relative ease of production of enzymes from bacteria or fungi encouraged the use of protein engineering to modify their substrate specificity, thermostability and enantioselectivity for biocatalysis applications. These proteins are used in various processes ranging from the selective isolation of enantiomeric precursors for biomedical applications to industrial products, such as additives to detergents or decontaminants. This adaptability is a longstanding story for families such as the lipases used in washing agents for decades, but this may prove useful for many other subfamilies as recently exemplified by efficient transformation of butyryl-cholinesterase into a cocaine esterase (16). Furthermore, the natural substrate promiscuity displayed by members of the superfamily can now be exploited (17–20). However, to be efficient, these engineering techniques need information about active sites and the range of sequence and structure variability found in nature. In combining classifications and tools to compare available data on α/β-hydrolases, ESTHER may provide the bases for most of these requirements. Herein, we describe the main entry point of the database, that is, the Overall Table, and show how Active Site search and Family Tree building programs are implemented to classify families.
OVERALL TABLE AND FAMILY TREE
In the Pfam database, which groups most α/β-hydrolases, the AB_Hydrolase clan (CL0028) is subdivided into 66 families (21). The ESTHER database contains far more families. The number increased from 69 in 2004 to 89 in 2008 to now reach 148 families, some of them being grouped in 47 superfamilies. This large number of families results from the following two features: firstly, because we subdivided some large families according to sequence similarities and biological information available from seed sequences; secondly, because we included some new families in the time intervals between major Pfam releases. Pfam, InterPro (22) or Uniprot (23) databases are not always fully convergent, and we often use the subdivision of the database that uses the most detailed subdivision. Some of the divisions are specific to ESTHER. In the new Overall Table, totally redesigned compared with the original version, the families are organized into three levels of grouping, so that there are blocks and families of rank number 1 and rank number 2. These subdivisions already give an idea of relationships between families. The Overall Table contains not only links to other general database resources but also buttons with pop-up windows showing taxonomic coverage, mutations, structures and substrates. Other buttons lead to specialized databases or classifications as described later in the text. To make it easier to compare protein functions in different families, we designed a method to built a Family Tree based on Hidden Markov Model (HMM) comparison (11). The Tree can be visualized, and it is fully interactive.
CORRESPONDENCE AND OVERLAP WITH OTHER ENZYME CLASSIFICATIONS OR DATABASES
The families described in ESTHER have corresponding entries in major general classification databases (PIR, InterPro, Pfam) and Proteopedia (24) and in Wikipedia when annotated entries are sufficiently curated. A few other specialized databases or published classifications also overlap with the classification of ESTHER. All of them have their specific purpose and serve different communities of biologists. In ESTHER, the Overall Table now provides links to these databases and a page summarizes all the correspondences.
Bacterial lipases
The first bacterial lipases to be recognized as members of the superfamily were enzymes homologous to the mammalian hormone-sensitive lipases (25,26). Later on, bacterial lipases and esterases were ordered in eight families (27). Since then, this classification has been widely used and extended (28–38), although it has not been integrated in any web server or database. We introduced this classification in ESTHER (Table 1). Among the bacterial lipases, six of the eight families originally described are related to α/β-hydrolases, but not the other two, families II and VIII, which belong to the SGNH-hydrolase and β-lactamase folds, respectively. It is noteworthy that in many and even small families, bacterial enzymes have eukaryotic homologues. The classification is extended, for example, family V has been subdivided in three subfamilies according to ESTHER families.
Table 1.
Correspondence of ESTHER families with the Arpigny and Jaeger classification
| ESTHER families | Arpigny and Jaeger families |
|---|---|
| Bacterial_lipase | Family_I |
| Bacterial_lip_FamI.1 | Family I.1 |
| Bacterial_lip_FamI.2 | Family_I.2 |
| Bacterial_lip_FamI.3 | Family_I.3 |
| Lipase_2 | Family_I.4 |
| Bacterial_lip_FamI.5 | Family_I.5 |
| Bacterial_lip_FamI.6 | Family_I.6 |
| Lipase_2 | Family_I.7 |
| Not α/β-hydrolase SGNH | Family_II |
| Polyesterase-lipase-cutinase | Family_III |
| Hormone-sensitive_lipase_like | Family_IV (25,26) |
| Hormone-sensitive_lipase_like_1 | Family_IV |
| ABHD6-Lip | Family_V.1 |
| Carboxymethylbutenolide_lactonase | Family_V.2 |
| UCP031982 | Family_V.3 (30) |
| LYsophospholipase_carboxylesterase | Family_VI |
| Carb_B_Bacteria | Family_VII |
| (not α/β-hydrolase β-lactamase) | Family_VIII |
| PHAZ7_phb_depolymerase | Family_IX (31) |
| Bacterial_EstLip_FamX | Family_X.1 (32) |
| Fungal-Bact_LIP | Family_X.2 (33) |
| Lipase_3 | Family_XI (34) |
| Bact_LipEH166_FamXII | Family_XII (35) |
| CarbLipBact | Family_XIII (32) |
| PC-sterol_acyltransferase | Family_XIV (36) |
| Duf_3089 | Family_XV (37,38) |
| Bacterial_Est97 | Family_XVI (39) |
Carbohydrate esterases
The CAZy database (Carbohydrate-Active Enzymes Database: http://www.cazy.org) describes the families of structurally related enzymes that degrade, modify or create glycosidic bonds (39). Among those, 6 families of 16 correspond to proteins belonging to the α/β-hydrolase fold (Table 2). The others belong to diverse folds, (β/α)7 barrel, (β)-helix, (β/α)8 barrel, 2-layer-sandwich, PIG-L, SGNH-hydrolase. At least in one family (CE10), the vast majority (if not all) of the members are esterases acting on non-carbohydrate substrates. The information on this family is no longer updated in CAZy, but in ESTHER, it represents Block C, which contains some of the most populated families.
Table 2.
Correspondence of ESTHER families with the CAZy classification of carbohydrate hydrolases
| Families in ESTHER | Families in CAZy |
|---|---|
| Antigen85c | CE-1 |
| Acetylxylan_esterase | CE-5 |
| Cutinase | CE-5 |
| Acetyl-esterase_deacetylase | CE-7 |
| Carb_B_Bacteria | CE-10 |
| Pectinacetylesterase-Notum | CE-13 |
| Glucuronoyl-esterase | CE-15 |
| Folds of families which are not α/β-hydrolases | |
| (β/α)7 barrel | CE-4 |
| (β)-helix | CE-8 |
| (β/α)8 barrel | CE-9 |
| 2-layer-sandwich | CE-11 |
| PIG-L | CE-14 |
| SGNH | CE-2, CE-3, CE-6, CE-12, CE-16 |
Serine peptidases
The MEROPS database (http://merops.sanger.ac.uk/) is an information resource for proteolytic enzymes acting on peptide bonds. Each clan is identified with two letters, of which the first represents the catalytic type of the families included in the clan (40) and the second is an arbitrary second capital letter. Among the 12 clans of serine peptidases, only one clan (SC) contains α/β-hydrolases. Yet, this clan is one of the most populated clans, with six families (S9 prolyl oligopeptidase, S10 carboxypeptidase Y, S15 Xaa-Pro dipeptidyl-peptidase, S28 lysosomal Pro-Xaa carboxypeptidase, S33 prolyl aminopeptidase, S37 PS-10 peptidase) that are all α/β-hydrolase families (Table 3).
Table 3.
Correspondence of peptidase families in ESTHER with clans and families in MEROPS
| Families in ESTHER | Families in MEROPS |
|---|---|
| Peptidase_S9 | S9 prolyl oligopeptidase |
| Prolyl_oligopeptidase_S9 | S9A |
| S9N_PPCE_Peptidase_S9 | S9A |
| S9N_PREPL_Peptidase_S9 | S9A |
| ACPH_Peptidase_S9 | S9C |
| DPP4N_Peptidase_S9 | S9B |
| Glutamyl_Peptidase_S9 | S9D |
| PMH_Peptidase_S9 | S9_upw |
| Carboxypeptidase_S10 | S10 carboxypeptidase Y |
| Peptidase_S15 | S15 Xaa-Pro dipeptidyl-peptidase |
| Cocaine_esterase | S15 |
| Lactobacillus_peptidase | S15 |
| Prolylcarboxypeptidase | S28 lysosomal Pro-Xaa carboxypeptidase |
| Proline_iminopeptidase | S33 prolyl aminopeptidase |
| Peptidase_S37 | S37 PS-10 peptidase |
Thioesterases
ThYme (Thioester-Active Enzyme Database: http://www.enzyme.cbirc.iastate.edu) is dedicated to enzymes involved in fatty acid and polyketide synthesis along with enzymes acting on thioester-containing substrates (41). Among the nine enzyme groups, the Thioesterase group contains 8 of 25 families that belong to the α/β-hydrolase fold and, as such, have counterpart entries in ESTHER (Table 5). The correspondence between the ESTHER and ThYme families was presented earlier (42). We extended the Overall Table and introduced the corresponding links into ESTHER.
Table 5.
Correspondence of ESTHER families with the thioesterase families in ThYme
| Families in ESTHER | Families in ThYme |
|---|---|
| Acyl-CoA_Thioesterase | TE-2
|
| Thioesterase | TE-16
|
| Thioesterase | TE-17 domain of polyketide synthase (PKS) |
| Thioesterase | TE-18
|
| Thioesterase_acyl-transferase | TE-19 luxD |
| Palmitoyl-protein_thioesterase | TE-20 Palmitoyl-protein thioesterase (ppt1, ppt2) |
| Lysophospholipase_carboxylesterase | TE-21
|
| A85-EsteraseD-FGH | TE-22
|
Lipases
LED (Lipase Engineering Database: http://www.led.uni-stuttgart.de/) is an internet database, which integrates information on sequence and structure of lipases and related proteins sharing the same α/β-hydrolase fold to facilitate protein engineering (43). Although this database overlaps with many ESTHER families, no correspondences were available between these classifications. Table 4 now fulfills this need, and the corresponding links have been introduced into ESTHER.
Table 4.
Correspondence of ESTHER families with the classification in LED
| Families in ESTHER | Families in LED |
|---|---|
| Block C and H | Class GGGX |
| Carboxylesterase | abH01 - Carboxylesterase |
| Fungal_carboxylesterase_lipase | abH02 - Yarrowia lipolytica lipase like - 03 |
| Fungal_carboxylesterase_lipase | abH03 - Candida rugosa lipase like |
| Hormone-sensitive_lipase_like | abH04 - Moraxella lipase 2 like |
| Hormone-sensitive_lipase_like | abH05 - Hormone sensitive lipases |
| Hormone-sensitive_lipase_like | abH06 - Brefeldin A esterase like |
| Class Y | |
| DPPIV_Peptidase S9 | abH27 - Dipeptidyl peptidase IV like |
| Prolylendopeptidase | abH28 - Prolyl endopeptidases |
| Lactobacillus_peptidase | abH29 - Dipeptidyl-peptidases |
| Cocaine_esterase | abH30 - Cocaine esterases |
| Fungal-Bact_LIP | abH38 - Candida antarctica lipase A like |
| Class GX | |
| Lipase_3 | abH23 - Filamentous fungi lipases |
| Thioesterase | abH07 - Moraxella lipase 3 like |
| Lipase_2 | abH18 - Bacillus lipases |
| Bacterial_lipase | abH15 - Burkholderia lipases |
| Lipase_3 | abH37 - Candida antarctica lipase like |
| Bacterial_Lipase | abH24 - Pseudomonas lipases |
| Acidic_lipase | abH14 - Gastric lipases |
| Pancreatic_lipase | abH20 - Lipoprotein lipases |
| Cutinase | abH36 - Cutinases |
| Polyesterase-lipase-cutinase | abH25 - Moraxella lipase 1 like |
| Epoxyde_hydrolase Proline imminopeptidase | abH09 - Microsomal Hydrolases |
| Epoxide hydrolase Haloalkane dehalogenase Non-heme peroxydase Carbon-carbon_bond_hydrolase CIB-CCG1-interacting-factor-B | abH08 - Cytosolic Hydrolases |
| Thioesterase_acyl-transferase | abH35 - Acyl-transferases |
| Lysophospholipase_carboxylesterase | abH22 - Lysophospholipase |
| Lipase_2 | abH16 - Streptomyces lipases |
| CarbLipBact | abH11 - Carboxylesterases |
| LYsophospholipase_carboxylesterase | abH21 - Bacterial esterases |
| Not α/β-hydrolase | abH32 - Xylanase esterases |
| Antigen85c | abH33 - Antigen 85 |
| Carboxypeptidase | abH34 - Lysosomal protective protein like |
| Acetyl-esterase_deacetylase | abH26 - Deacetylases |
| Bacterial_esterase | abH13 - Bacterial esterases |
| Hydroxynitrile_lyase | abH12 - Hydroxynitrile lyases |
| Palmitoyl-protein_thioesterase | abH19 - Thioesterases |
| Dienelactone_hydrolase | abH31 - Dienlactone Hydrolases |
| PGAP1 | abH17 - Chloroflexus aurantiacus lipase-like |
| AlphaBeta_hydrolase | abH10 - Uncultured crenarchaeote |
VARIATION IN CORE ARCHITECTURE
An important variation of the canonical core architecture was revealed by the crystal structure of Penicillium finiculosum polyhydroxybutyrate depolymerase, where the sequences dictating β-strands 1–4 in the central β-sheet are circularly permuted from the N- to the C-terminal of the protein (44). In the patatin family, which is closely related to the α/β-hydrolase fold family (45), a recent crystal structure of the acyltransferase domain of the iterative polyketide synthase (DynE8) from Micromonospora chersina shows an even more drastic split of the β-sheet core (46). Indeed, the first four β-strands are separated from the rest of the β-sheet and serve as a linker between the ketoacyl synthase and acyltransferase domains of the protein. Such a split of the central β-sheet has not been described in α/β-hydrolases, but it might occur for members known only by their genomic sequence where large insertions are found between predicted β-strands.
VARIATION IN PREDICTED ACTIVE SITE RESIDUES
The conservation degree of amino acid residues in individual sequences against the aligned sequences of the family is now visualized in ESTHER with a colour code; the darker the shade, the higher the conservation. The putative active site residues are also highlighted when this information can be derived from the best-characterized homologues within the family.
The most common active site nucleophilic residue in α/β-hydrolase enzymes is a Ser located within a GXSXG pentapeptide (where X denotes any residue), whereas the other two active site residues are an acidic residue (Glu or Asp) and a His as a general acid–base catalyst. In dienelactone hydrolases, the nucleophilic residue is a Cys, and in haloalkane dehalogenases, it is an Asp. In glycoside hydrolases, there are nearly always two catalytic residues, both either Asp or Glu. Recently, in E. coli RutD a His has been found to substitute the canonical Ser, to form a GHALG pentapeptide in the nucleophile elbow (47). Even more surprising, in tomato methyl ketone synthase 1 (MKS1), a 3-ketoacid decarboxylase that clearly clusters in a subclass of the hydroxynitrile lyase family, the nucleophilic residue is Ala87, central to the GHALG pentapeptide. This substitution seems to have occurred only recently in the Lycopodium lineage, as many close homologues from plants retain a Ser in their active site (48). Yet, the Ala87Ser mutant in MKS1 displays greater decarboxylation activity than the wild-type enzyme, arguing for a limited evolutionary history within a functionally constrained and structurally ‘highly conserved’ active site region. In all the active sites analysed, the only conserved residue is the His. However, this residue generally lies within a sequence that is not particularly well conserved between families, whereas it is often conserved in non-catalytic proteins. Hence, it cannot serve as a marker for inferring that a protein is an enzymatically active α/β-hydrolase.
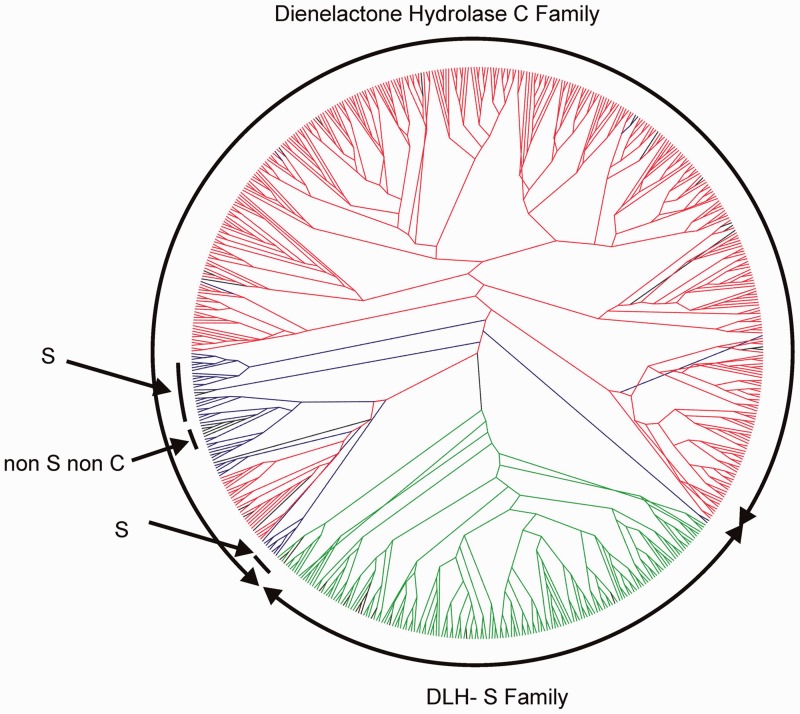
However, even though extracting information from structural data for inferring the catalytic residues in all members of families remains arduous, we attempted to do so with the aim of improving the subdivision of families. For example, we used the occurrence of an Asp or a Glu as the putative active-site acidic residue to split the large family called Antigen85c in ESTHER and PF00756 Esterase in Pfam. As a result, this family is now separated in five subfamilies, three have an Asp (A85-EsteraseD-FGH, A85-Feruloyl-Esterase, A85-Est-Putative) and two have a Glu (A85-IroE-IroD-Fes-Yiel, A85-Mycolyl-transferase). Another example is diene-lactone hydrolase, whose active-site nucleophilic residue is a Cys; yet, a recent crystal structure of a related Anabaena variabilis enzyme shows a Ser in the homologous position (Protein Data Bank code, 2O2G). This sequence is annotated as a dienelactone hydrolase, and many sequences issued from whole genome sequencing are annotated accordingly. We analysed together sequences from the original dienelactone hydrolase family and sequences homologous to the newly annotated sequence and built a Family Tree from the aligned sequences (Figure 1). This procedure led us to clearly separate a family, which we called DLH-S (with S for Ser). Although members of this family are annotated dienelactone hydrolases, too little data are available for inferring a clear function for this new family. The Tree also shows some sequences truly homologous to well characterized dienelactone hydrolases that have a Ser in the active site, or groups of sequences that have neither Ser nor Cys in homologous position of the active-site nucleophilic residue. Overall, this kind of analysis can help clarify the strategy and results from genome annotation and sequence analysis. We plan to refine, extend and standardize this approach to other ESTHER families of in the near future.
Figure 1.
Sequences of putative dienelactone hydrolases where aligned with Clustal Omega (49). The Family Tree was built with FastTree 2 (50), and branches were coloured according to the nature of the nucleophilic residue in the active site. Two families were separated, in blue and red the Dienelactone_hydrolase family and in green the DLH-S family. Blue branches are sequences that have a Cys suitably positioned to be the nucleophilic active site residue, whereas green and red branches have a Ser. Black branches are sequences that have neither Cys nor Ser.
FUNDING
Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 2.Nardini M, Dijkstra BW. α/β hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 3.Heikinheimo P, Goldman A, Jeffries C, Ollis DL. Of barn owls and bankers: a lush variety of α/β hydrolases. Structure. 1999;7:141–146. doi: 10.1016/s0969-2126(99)80079-3. [DOI] [PubMed] [Google Scholar]
- 4.Holmquist M. Alpha/beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 5.Carr PD, Ollis DL. α/β hydrolase fold: an update. Protein Pept. Lett. 2009;16:1137–1148. doi: 10.2174/092986609789071298. [DOI] [PubMed] [Google Scholar]
- 6.Cousin X, Hotelier T, Lievin P, Toutant JP, Chatonnet A. A cholinesterase genes server (ESTHER): a database of cholinesterase-related sequences for multiple alignments, phylogenetic relationships, mutations and structural data retrieval. Nucleic Acids Res. 1996;24:132–136. doi: 10.1093/nar/24.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cousin X, Hotelier T, Giles K, Lievin P, Toutant JP, Chatonnet A. The α/β fold family of proteins database and the cholinesterase gene server ESTHER. Nucleic Acids Res. 1997;25:143–146. doi: 10.1093/nar/25.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cousin X, Hotelier T, Giles K, Toutant JP, Chatonnet A. aCHEdb: the database system for ESTHER, the α/β fold family of proteins and the Cholinesterase gene server. Nucleic Acids Res. 1998;26:226–228. doi: 10.1093/nar/26.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatonnet A, Cousin X, Robinson A. Links between kinetic data and sequences in the alpha/beta-hydrolases fold database. Brief Bioinform. 2001;2:30–37. doi: 10.1093/bib/2.1.30. [DOI] [PubMed] [Google Scholar]
- 10.Renault L, Nègre V, Hotelier T, Cousin X, Marchot P, Chatonnet A. New friendly tools for users of ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Chem. Biol. Interact. 2005;157–158:339–343. doi: 10.1016/j.cbi.2005.10.100. [DOI] [PubMed] [Google Scholar]
- 11.Lenfant N, Hotelier T, Bourne Y, Marchot P, Chatonnet A. Proteins with an alpha/beta hydrolase fold: relationships between subfamilies in an ever-growing superfamily. Chem. Biol. Interact. 2012 doi: 10.1016/j.cbi.2012.09.003. pii: S0009-2797(12)00168-8 September 23 (doi:10.1016/j.cbi.2012.09.003; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 12.Hotelier T, Renault L, Cousin X, Nègre V, Marchot P, Chatonnet A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Nucleic Acids Res. 2004;32:D145–D147. doi: 10.1093/nar/gkh141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotelier T, Nègre V, Marchot P, Chatonnet A. Insecticide resistance through mutations in cholinesterases or carboxylesterases: data mining in the ESTHER database. J. Pestic. Sci. 2010;35:315–320. [Google Scholar]
- 14.Bugg TD. Diverse catalytic activities in the α/β-hydrolase family of enzymes: activation of H2O, HCN, H2O2, and O2. Bioorg. Chem. 2004;32:367–375. doi: 10.1016/j.bioorg.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Marchot P, Chatonnet A. Enzymatic activity and protein interactions in alpha/beta hydrolase fold proteins: moonlighting versus promiscuity. Protein Pept. Lett. 2012;19:132–143. doi: 10.2174/092986612799080284. [DOI] [PubMed] [Google Scholar]
- 16.Xue L, Hou S, Yang W, Fang L, Zheng F, Zhan CG. Catalytic activities of a cocaine hydrolase engineered from human butyrylcholinesterase against (+)- and (-)-cocaine. Chem. Biol. Interact. 2012 doi: 10.1016/j.cbi.2012.08.003. August 11 (doi:10.1016/j.cbi.2012.08.003; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawanami T, Miyakoshi M, Dairi T, Itoh N. Reaction mechanism of the Co2+-activated multifunctional bromoperoxidase-esterase from Pseudomonas putida IF-3. Arch. Biochem. Biophys. 2002;398:94–100. doi: 10.1006/abbi.2001.2702. [DOI] [PubMed] [Google Scholar]
- 18.Li C, Hassler M, Bugg TD. Catalytic promiscuity in the α/β-hydrolase superfamily: hydroxamic acid formation, C–C bond formation, ester and thioester hydrolysis in the C–C hydrolase family. Chembiochem. 2008;9:71–76. doi: 10.1002/cbic.200700428. [DOI] [PubMed] [Google Scholar]
- 19.Thierbach S, Büldt-Karentzopoulos K, Dreiling A, Hennecke U, König S, Fetzner S. Hydrolase-like properties of a cofactor-independent dioxygenase. Chembiochem. 2012;13:1125–1127. doi: 10.1002/cbic.201200152. [DOI] [PubMed] [Google Scholar]
- 20.Manco G, Merone L, Porzio E, Feng Y, Mandrich L. Enzyme promiscuity in the hormone-sensitive lipase family of proteins. Protein Pept. Lett. 2012;19:144–154. doi: 10.2174/092986612799080400. [DOI] [PubMed] [Google Scholar]
- 21.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prilusky J, Hodis E, Canner D, Decatur WA, Oberholser K, Martz E, Berchanski A, Harel M, Sussman JL. Proteopedia: a status report on the collaborative, 3D web-encyclopedia of proteins and other biomolecules. J. Struct. Biol. 2011;175:244–252. doi: 10.1016/j.jsb.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Langin D, Holm C. Sequence similarities between hormone-sensitive lipase and five prokaryotic enzymes. Trends Biochem. Sci. 1993;18:466–467. doi: 10.1016/0968-0004(93)90007-a. [DOI] [PubMed] [Google Scholar]
- 26.Hemila H, Koivula TT, Palva I. Hormone-sensitive lipase is closely related to several bacterial proteins, and distantly related to acetylcholinesterase and lipoprotein lipase: identification of a superfamily of esterases and lipases. Biochim. Biophys. Acta. 1994;1210:249–253. doi: 10.1016/0005-2760(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 27.Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- 28.Jaeger KE, Eggert T. Lipases for biotechnology. Curr. Opin. Biotechnol. 2002;13:390–397. doi: 10.1016/s0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- 29.Rao L, Xue Y, Zhou C, Tao J, Li G, Lu JR, Ma Y. A thermostable esterase from Thermoanaerobacter tengcongensis opening up a new family of bacterial lipolytic enzymes. Biochim. Biophys. Acta. 2011;1814:1695–1702. doi: 10.1016/j.bbapap.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Park SY, Kim JT, Kang SG, Woo JH, Lee JH, Choi HT, Kim SJ. A new esterase showing similarity to putative dienelactone hydrolase from a strict marine bacterium, Vibrio sp. GMD509. Appl. Microbiol. Biotechnol. 2007;77:107–115. doi: 10.1007/s00253-007-1134-2. [DOI] [PubMed] [Google Scholar]
- 31.Handrick R, Reinhardt S, Focarete ML, Scandola M, Adamus G, Kowalczuk M, Jendrossek D. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J. Biol. Chem. 2001;276:36215–36224. doi: 10.1074/jbc.M101106200. [DOI] [PubMed] [Google Scholar]
- 32.Levisson M, Van der Oost J, Kengen SW. Characterization and structural modeling of a new type of thermostable esterase from Thermotoga maritima. FEBS J. 2007;274:2832–2842. doi: 10.1111/j.1742-4658.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 33.Bassegoda A, Pastor FI, Diaz P. Rhodococcus sp. strain CR-53 LipR, the first member of a new bacterial lipase family (family X) displaying an unusual Y-type oxyanion hole, similar to the Candida antarctica lipase clan. Appl. Environ. Microbiol. 2012;78:1724–1732. doi: 10.1128/AEM.06332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee MH, Lee CH, Oh TK, Song JK, Yoon JH. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl. Environ. Microbiol. 2006;72:7406–7409. doi: 10.1128/AEM.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim EY, Oh KH, Lee MH, Kang CH, Oh TK, Yoon JH. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl. Environ. Microbiol. 2009;75:257–260. doi: 10.1128/AEM.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee MH, Hong KS, Malhotra S, Park JH, Hwang EC, Choi HK, Kim YS, Tao W, Lee SW. A new esterase EstD2 isolated from plant rhizosphere soil metagenome. Appl. Microbiol. Biotechnol. 2010;88:1125–1134. doi: 10.1007/s00253-010-2729-6. [DOI] [PubMed] [Google Scholar]
- 37.Bayer S, Kunert A, Ballschmiter M, Greiner-Stoeffele T. Indication for a new lipolytic enzyme family: isolation and characterization of two esterases from a metagenomic library. J. Mol. Microbiol. Biotechnol. 2010;18:181–187. doi: 10.1159/000315459. [DOI] [PubMed] [Google Scholar]
- 38.Fu J, Leiros HK, de Pascale D, Johnson KA, Blencke HM, Landfald B. Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol. 2012 doi: 10.1007/s00253-012-4276-9. July 26 (doi:10.1007/s00253-012-4276-9; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 39.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantu DC, Chen Y, Reilly PJ. Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cantu DC, Chen Y, Lemons ML, Reilly PJ. ThYme: a database for thioester-active enzymes. Nucleic Acids Res. 2011;39:D342–D346. doi: 10.1093/nar/gkq1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer M, Pleiss J. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acids Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hisano T, Kasuya KI, Tezuka Y, Ishii N, Kobayashi T, Shiraki M, Oroudjev E, Hansma H, Iwata T, Doi Y, et al. The crystal structure of polyhydroxybutyrate depolymerase from Penicillium funiculosum provides insights into the recognition and degradation of biopolyesters. J. Mol. Biol. 2005;356:993–1004. doi: 10.1016/j.jmb.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 45.Kienesberger PC, Oberer M, Lass A, Zechner R. Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions. J. Lipid. Res. 2009;50(Suppl.):S63–S68. doi: 10.1194/jlr.R800082-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liew CW, Nilsson M, Chen MW, Sun H, Cornvik T, Liang ZX, Lescar J. Crystal structure of the acyltransferase domain of the iterative polyketide synthase in enediyne biosynthesis. J. Biol. Chem. 2012;287:23203–23215. doi: 10.1074/jbc.M112.362210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knapik AA, Petkowski JJ, Otwinowski Z, Cymborowski MT, Cooper DR, Majorek KA, Chruszcz M, Krajewska WM, Minor W. A multi-faceted analysis of RutD reveals a novel family of α/β hydrolases. Proteins. 2012;80:2359–2368. doi: 10.1002/prot.24122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auldridge ME, Guo Y, Austin MB, Ramsey J, Fridman E, Pichersky E, Noel JP. Emergent decarboxylase activity and attenuation of α/β-hydrolase activity during the evolution of methylketone biosynthesis in tomato. Plant Cell. 2012;24:1596–1607. doi: 10.1105/tpc.111.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Systems Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan N, Price MN, Dehal PS, Arkin AP. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]