Abstract
The University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu) offers online public access to a growing database of genomic sequence and annotations for a wide variety of organisms. The Browser is an integrated tool set for visualizing, comparing, analysing and sharing both publicly available and user-generated genomic datasets. As of September 2012, genomic sequence and a basic set of annotation ‘tracks’ are provided for 63 organisms, including 26 mammals, 13 non-mammal vertebrates, 3 invertebrate deuterostomes, 13 insects, 6 worms, yeast and sea hare. In the past year 19 new genome assemblies have been added, and we anticipate releasing another 28 in early 2013. Further, a large number of annotation tracks have been either added, updated by contributors or remapped to the latest human reference genome. Among these are an updated UCSC Genes track for human and mouse assemblies. We have also introduced several features to improve usability, including new navigation menus. This article provides an update to the UCSC Genome Browser database, which has been previously featured in the Database issue of this journal.
INTRODUCTION
The University of California Santa Cruz (UCSC) Genome Browser (1,2) at http://genome.ucsc.edu is a web-based set of tools providing access to a database of genome sequence and annotations for visualization, comparison and analysis by the scientific, medical and academic communities. Our primary mission is to provide timely and convenient open access to high-quality human genome sequence and annotations in a framework that enables easy exploration from genome-wide down to the base level. Annotation datasets, or ‘tracks’, on the human genome cover conservation and evolutionary comparisons, gene models, regulation, expression, epigenetics and tissue differentiation, variation, phenotype and disease associations. Our mission extends to a number of additional organisms including 6 other primates, 19 additional mammals including 3 marsupials and 1 monotreme, 13 non-mammalian vertebrates and 24 invertebrates, each with varying degrees of genome-specific annotation. Many of the genomes in our database have multiple assembly versions, which support researchers who use annotations mapped using older assemblies.
LOCAL DATASETS
The Genome Browser locally hosts mapping and sequence annotation tracks that describe assembly, gap and GC content for all organisms in the browser database. Additionally, for most organisms we show alignments from RefSeq genes (3), mRNAs and ESTs from GenBank (4), and other gene or gene prediction tracks such as Ensembl Genes (5). For human and mouse assemblies, we also offer a locally generated UCSC Genes track based upon RefSeq, GenBank, CCDS and UniProt data (6,7). About half of the genomes hosted at UCSC include a multiple sequence alignment (multiz) track (8) and pairwise genomic alignments between assemblies to facilitate comparative and evolutionary investigations. Expression, regulation, variation and phenotype tracks are available for many of the assemblies. Most locally hosted tracks include descriptions with references and links to the original contributors or research upon which the annotations are based.
New genome assemblies
With the abundance of new vertebrate assemblies available in GenBank, the UCSC Genome Browser team has streamlined its browser release pipeline in the effort to keep pace. We have added 19 new assemblies to the Genome Browser in the past year, including 4 model organisms (Fugu, mouse, worm and yeast), 7 newly sequenced organisms (gibbon, lesser hedgehog tenrec, medium ground finch, naked mole-rat, tasmanian devil, turkey and western painted turtle) and 8 updated assemblies for previously published organisms (chicken, cow, dog, gorilla, microbat, rat, tammar wallaby and western clawed frog)—see Table 1 for details. We anticipate the public release of 28 more genome assemblies in the coming months (Table 2) in support of the new mouse (GRCm38/mm10) 60-way conservation track. For a complete list of the genome assemblies included in this track, refer to the mm10 Conservation track description page on the Genome Browser website.
Table 1.
Assemblies released on the Genome Browser in 2012
| Common name | Scientific name | UCSC ID | Sequencing center | Sequencing center ID | Notes |
|---|---|---|---|---|---|
| Chicken | Gallus gallus | galGal4 | Int’l Chicken GSC | Gallus_gallus-4.0 | |
| Cow | Bos Taurus | bosTau7 | Cattle GSC | Btau_4.6.1 | |
| Dog | Canis familiaris | canFam3 | Dog GSC | V3.1 | |
| Fugu | Takifugu rubripes | fr3 | Int’l Fugu GSC | FUGU5 | RefSeq Genes, 8-species mult. alignment |
| Gibbon | Nomascus leucogenys | nomLeu1 | Gibbon GSC | Nleu1.0 | |
| Gorilla | Gorilla gorilla gorilla | gorGor3 | Wellcome Trust Sanger Institute | gorGor3.1 | |
| Lesser hedgehog tenrec | Echinops telfairi | echTel1 | Broad Institute | EchTel1 | |
| Medium ground finch | Geospiza fortis | geoFor1 | Genome 10K Project and BGI | GeoFor_1.0 | |
| Microbat | Myotis lucifugus | myoLuc2 | Broad Institute | Myoluc2.0 | |
| Mouse | Mus musculus | mm10 | Mouse GRC | GRCm38 | RefSeq Genes, 60-species mult. alignment |
| Naked mole-rat | Heterocephalus glaber | hetGla1 | BGI | HetGla_1.0 | |
| Rat | Rattus | rn4 | Baylor Human GSC | RGSC_v3.4 | |
| Tammar wallaby | Macropus eugenii | macEug2 | Tammar Wallaby GSC | Meug_1.1 | |
| Tasmanian devil | Sarcophilus harrisii | sarHar1 | Wellcome Trust Sanger Institute | Devil_refv7.0 | |
| Turkey | Meleagris gallopavo | melGal1 | Turkey GSC | Turkey_2.01 | |
| Western clawed frog | Xenopus (Silurana) tropicalis | xenTro3 | US DOE JGI-PGF | V4.2 | |
| Western painted turtle | Chrysemys picta bellii | chrPic1 | Int’l Painted Turtle GSC | Chrysemys_picta_bellii-3.0.1 | |
| Worm | Caenorhabditis elegans | ce10 | WormBase | WS220 | RefSeq Genes, 7-species mult. alignment |
| Yeast | Saccharomyces cerevisiae | sacCer3 | Saccharomyces Genome Database (SGD) | SacCer_Apr2011 | Ensembl Genes, 7-species mult. alignment |
Table 2.
Assemblies to be released on the Genome Browser by early 2013
| Common name | Scientific name | UCSC ID | Sequencing center | Sequencing center ID |
|---|---|---|---|---|
| Alpaca | Vicugna pacos | vicPac1 | Broad Institute | VicPac1.0 |
| Armadillo | Dasypus novemcinctus | dasNov3 | Baylor College of Medicine (BCM) | Dasnov3.0 |
| Atlantic cod | Gadus morhua | gadMor1 | Genofisk | GadMor_May2010 |
| Baboon | Papio hamadryas | papHam1 | BCM | Pham_1.0 |
| Budgerigar | Melopsittacus undulates | melUnd1 | Washington University at St. Louis | Melopsittacus_undulatus_6.3 |
| Bushbaby | Otolemur garnettii | otoGar3 | Broad Institute | OtoGar3 |
| Cat | Felis catus | felCat5 | International Cat GSC | Felis_catus 6.2 |
| Chimpanzee | Pan troglodytes | panTro4 | Chimpanzee SAC | CSAC 2.1.4 |
| Chinese rhesus | Macaca mulatta | rheMac3 | BGI | CR_1.0 |
| Coelacanth | Latimeria chalumnae | latCha1 | Broad Institute | LatCha1 |
| Dolphin | Tursiops truncates | turTru2 | BCM | Ttru_1.4 |
| Gibbon | Nomascus leucogenys | nomLeu2 | Gibbon GSC | Nleu1.1 |
| Hedgehog | Erinaceus europaeus | eriEur1 | Broad Institute | EriEur1 |
| Kangaroo rat | Dipodomys ordii | dipOrd1 | Broad Institute | DipOrd1.0 |
| Manatee | Trichechus manatus latirostris | triMan1 | Broad Institute | TriManLat1.0 |
| Megabat | Pteropus vampyrus | pteVam1 | Broad Institute | PteVap1.0 |
| Mouse lemur | Microcebus murinus | micMur1 | Broad Institute | MicMur1.0 |
| Naked mole-rat | Heterocephalus glaber | hetGla2 | Broad Institute | HetGla_female_1.0 |
| Nile tilapia | Oreochromis niloticus | oreNil1 | Broad Institute | Orenil1.0 |
| Pig | Sus scrofa | susScr3 | International Swine GSC | Sscrofa10.2 |
| Pika | Ochotona princeps | ochPri2 | Broad Institute | OchPri2.0 |
| Rock hyrax | Procavia capensis | proCap1 | Broad Institute | ProCap1.0 |
| Shrew | Sorex araneus | sorAra1 | Broad Institute | SorAra1 |
| Sloth | Choloepus hoffmanni | choHof1 | Broad Institute | ChoHof1.0 |
| Squirrel | Spermophilus tridecemlineatus | speTri2 | Broad Institute | SpeTri2.0 |
| Squirrel monkey | Saimiri boliviensis | saiBol1 | Broad Institute | SaiBol1.0 |
| Tarsier | Tarsius syrichta | tarSyr1 | Broad Institute | TarSyr1.0 |
| Tree shrew | Tupaia belangeri | tupBel1 | Broad Institute | TupBel1 |
New and updated annotations
Many new datasets were added to the Genome Browser this year, and several existing datasets underwent major revisions. A significant portion of these were contributed by the Encyclopedia of DNA Elements (ENCODE) Consortium: we released tracks and downloadable files for more than 2300 experiments as the Data Coordination Center for the ENCODE Project (9,10), described in a companion paper in this issue.
We published a major update of the UCSC Genes track (6) for the human assembly (GRCh37/hg19) that includes more non-coding transcripts based on data from Rfam and from the tRNA Genes track. We anticipate releasing an updated UCSC Genes for mm10 in fall of 2012. Rat Genome Database (RGD) Genes for rat has replaced UCSC Genes as the main gene track for Baylor 3.4/rn4 (11).
We have updated dbSNP for hg19 to version 135, which includes interim phase 1 variant calls from the 1000 Genomes project (12). This new version contains additional annotation data not included in previous dbSNP tracks, with corresponding coloring and filtering options in the Genome Browser. We anticipate having dbSNP version 137 for hg19 available in fall 2012, with Sequence Ontology (13) terms replacing dbSNP's functional annotation terms in the display.
To ensure timely display of data from frequently updated phenotype and disease association databases we have automated loading of the following hg19 tracks: Catalogue Of Somatic Mutations In Cancer (COSMIC), GeneReviews, GWAS Catalog and Online Mendelian Inheritance in Man (OMIM) (14–17).
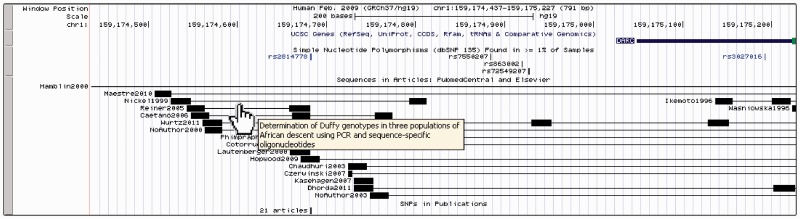
We have added a Publications track that shows DNA and protein sequences, SNPs, cytogenetic bands and gene symbols which were text-mined from 3 million biomedical articles in Elsevier, PubMed Central and other databases (18). This track is based on the UCSC Genocoding Project, which searches for references to chromosomal locations in scientific articles. The annotations in this track link back to the original article, thus allowing researchers to identify publications relevant to a particular locus (Figure 1).
Figure 1.
Genome Browser image of the promoter region of DARC on human assembly hg19 including UCSC Genes, dbSNP 135 and the Publications track showing sequences and SNPs text-mined from PubMed Central and Elsevier. The region shown includes a SNP responsible for the Duffy blood group (rs2814778). The publication track contains sequences in this region from several articles relevant to this SNP. Note that hovering the mouse over a sequence shows the title of the corresponding article. Clicking on a sequence in the publications track takes the user to a page with details about the relevant article.
We have added four public track hubs for hg19 from external data providers (see below for more details on track hubs): the ENCODE Analysis hub contains descriptions of ENCODE data in uniformly processed signal and element representations, as well as genome segmentations (19); the UMassMed ZHub contains H3K4me3 ChIP-seq data for autistic brains (20); the Expression & PolyA Database (xPAD) hub contains a map of polyadenylation sites in cancer tissues and tumor cell lines (21); the miRcode hub contains predicted microRNA target sites in GENCODE transcripts (22).
SOFTWARE IMPROVEMENTS
We made several changes to the interface of the Genome Browser in 2012 based on suggestions from our users. All pages now display a menu bar to make it easier to access features and navigate around the website in a consistent way. We have changed the fonts and background to improve usability. The annotation search and gene suggest box have been combined, and we have added descriptions to the gene suggestion list. We have changed the way users log in when saving sessions; this change simplifies the login procedure and also removes the dependency on MediaWiki, which makes it easier for Genome Browser mirrors to support saved sessions.
We introduced support for the Variant Call Format (VCF) in 2011 (23). This year we improved VCF support with a haplotype sorting display. VCF can optionally represent phased genotypes, i.e. the two alleles of each diploid genotype have been assigned to two haplotypes, one inherited from each parent. For VCF files that contain phased genotypes from multiple samples, we have developed an advanced display to highlight local patterns of genetic linkage between variants. The display features the clustering of independent haplotypes within the viewed region. The goal of the clustering is to visually group co-occurring allele sequences in haplotypes, so local patterns of linkage can be easily discerned. The clustering does not indicate relatedness of individuals, but merely local composition of mostly ancient haplotype blocks. We anticipate adding 1000 Genomes Phase 1 variant calls with phased genotypes for 1092 individuals using this display in fall 2012.
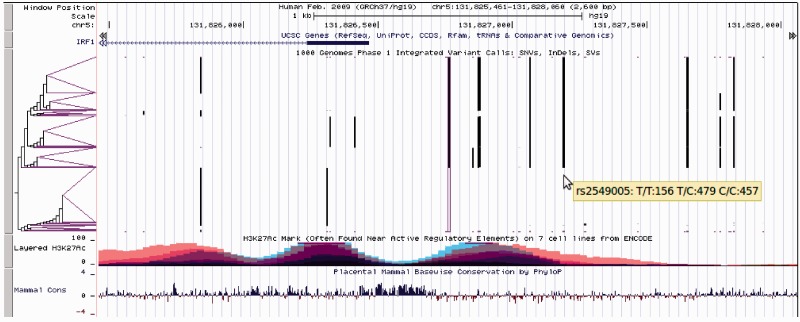
In the haplotype sorting display (Figure 2), independent haplotypes are shown horizontally, and variants are vertical bars with reference alleles in white (invisible) and alternate alleles in black. A variant for which most haplotypes have the reference allele will be mostly white (invisible); tick marks at the top and bottom of each variant make such variants easier to see. Haplotypes are clustered by similarity weighted by proximity to a central variant, which is outlined in purple. In order to limit compute time, only a small number of variants are used for clustering; these variants have purple tick marks above and below. The clustering tree is drawn in the left label area, and is used to order the haplotypes from top to bottom. When a rightmost branch in the clustering tree is purple, it means that all haplotypes in the branch are identical, at least in the variants used for clustering.
Figure 2.
Genome Browser image of the promoter region and transcription start of IRF1 on human assembly hg19 showing UCSC Genes, 1000 Genomes Phase 1 Integrated Variant Calls in the haplotype sorting VCF display mode, histone mark H3K27Ac binding in overlays of 7 ENCODE cell lines and PhyloP conservation scores from alignments of placental mammals. Mouse-over text gives the dbSNP identifier and genotype counts for one of the 1000 Genomes variants. The variant outlined in purple is used as the center variant for clustering haplotypes by similarity, and is clearly in linkage with nearby variants. Wider purple triangular leaves of the clustering tree indicate more common local haplotypes. Note that the reference genome haplotype (horizontal run of invisible reference alleles) is often not the major haplotype among the 1000 Genomes Phase 1 samples.
In 2011 we introduced support for track data hubs, which are web-accessible directories of genomic data that can be viewed in the UCSC Genome Browser alongside the annotation tracks hosted by UCSC (2). This technology has many advantages: it allows researchers to combine and configure large numbers of datasets for presentation as single entity, it improves performance by allowing the Genome Browser to retrieve data only when necessary, and it allows researchers to share a collection of data with colleagues as a private data hub. Track hubs usage increased greatly in 2012; by September 2012 more than 2000 track hubs were in use. There is also a growing trend in the research community to use track hubs to collect and organize data for presentation in publications. UCSC has extended the documentation (http://genome.ucsc.edu/goldenPath/help/trackDb/trackDbDoc.html) for track hubs on the Genome Browser website to facilitate their use.
FUTURE DIRECTIONS
We will continue to add new and updated genome assemblies for vertebrate and other selected model organisms as they become available. Only assemblies registered and deposited in NCBI’s GenBank will be considered for hosting at UCSC, as stipulated in the Browser Genome Release Agreement instituted by NCBI, Ensembl and UCSC. Many researchers have expressed interest in using the Genome Browser to visualize and analyse assemblies that are not deposited at NCBI. To assist such research, we intend to develop support for assembly data hubs, which will enable the genomics community to easily extend the Genome Browser to display genome assemblies that we are unable to integrate into our own database. The assembly data hub will be similar in concept to the track data hub: the data provider will store the genome sequence in a compressed, binary, indexed file format and make it available on a remote web server along with a list of tracks that annotate that genome.
We plan to add or update several annotation tracks in the upcoming year, including a coverage/mapability track based on 1000 Genomes project data, an updated recombination rate and UCSC Genes track for the human genome, an updated ORFeome track for zebrafish, a mouse strain variant track, segmental duplication tracks for several assemblies, and more selected personal genomes in the human Personal Genome Variants track. We will also continue to incorporate selected datasets from the ENCODE project that are of general interest to our users.
We are developing a tool for integrating diverse annotations in our databases with user-provided genomic variants, to assist with analysis and prioritization of variants discovered via sequencing. We will finish support for VCF in tracks hubs. We also plan to implement a supported mirror in Germany to improve access speed for European users of the Genome Browser.
CONTACTING US
We have two public, moderated mailing lists for user support: genome@soe.ucsc.edu for general questions about the Genome Browser and genome-mirror@soe.ucsc.edu for questions specific to the setup and maintenance of Genome Browser mirrors. Archives of both lists are searchable from our contacts page at http://genome.ucsc.edu/contacts.html. You may also reach us at genome-www@soe.ucsc.edu, the preferred address for inquiring about mirror site licenses and reporting server errors.
FUNDING
National Human Genome Research Institute [P41HG002371 to G.P.B., H.C., M.D., P.A.F., A.S.H., F.H., D.K., V.K., W.J.K., R.M.K., B.T.L., C.H.L., L.R.M, A.P., B.J.R., B.R., G.R. and A.S.Z.; U41HG004568 to M.S.C., T.R.D., M.G., F.H., W.J.K., K.L., V.S.M., B.J.R., K.R.R., C.A.S. and M.W.; and subcontracts from P01HG5062 to G.P.B., W.J.K. and B.R; U54HG004555 to M.D. and R.A.H.; U41HG004269 to A.S.H. and W.J.K.; U01HG004695 to W.J.K.]; subcontracts from the National Institute of Dental and Craniofacial Research [U01DE20057 to G.P.B. and R.M.K.]; National Institute of Child Health and Human Development [RC2HD064525 to H.C., A.S.H. and R.M.K.]; National Institute of Environmental Health Sciences [U01ES017154 to W.J.K]. European Molecular Biology Organization Long-Term Fellowship (ALTF 292-2011 to M.H.). Support from Howard Hughes Medical Institute (to D.H.). Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. G.P.B., H.C., M.D., T.R.D., P.A.F., B.M.G., D.H., R.A.H., A.S.H., D.K., V.K., W.J.K., R.M.K., K.L., C.H.L., V.S.M., L.R.M., A.P., B.R., B.J.R., K.R.R., C.A.S. and A.S.Z. receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities; W.J.K. works for Kent Informatics.
ACKNOWLEDGEMENTS
The authors would like to thank the many data contributors whose work makes the Genome Browser possible, our Scientific Advisory Board for steering our efforts, our users for their consistent support and valuable feedback, and our outstanding team of system administrators: Jorge Garcia, Erich Weiler and Gary Moro.
REFERENCES
- 1.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;18:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 7.Karolchik D, Kuhn R, Baertsch R, Barber G, Clawson H, Diekhans M, Giardine B, Harte R, Hinrichs A, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ, RGD Team. The Rat Genome Database, update 2007—easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherry S, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski E, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilbeck K, Lewis SE. Sequence Ontology annotation guide. Comp. Funct. Genomics. 2004;5:642–647. doi: 10.1002/cfg.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The catalogue of somatic mutations in cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008;57:10.11.1–10.11.26. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, Beahler C, Bird TD, Popovich B, Nesbitt C, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum. Mutat. 2002;19:501–509. doi: 10.1002/humu.10069. [DOI] [PubMed] [Google Scholar]
- 16.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's online Mendelian inheritance in man (OMIM®) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haeussler M, Gerner M, Bergman CM. Annotating genes and genomes with DNA sequences extracted from biomedical articles. Bioinformatics. 2011;27:980–986. doi: 10.1093/bioinformatics/btr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, Guo Y, Lessard A, Akbarian S, Weng Z. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch. Gen. Psychiatry. 2012;69:314–324. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40:8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCF tools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]