Abstract
ValidNESs (http://validness.ym.edu.tw/) is a new database for experimentally validated leucine-rich nuclear export signal (NES)-containing proteins. The therapeutic potential of the chromosomal region maintenance 1 (CRM1)-mediated nuclear export pathway and disease relevance of its cargo proteins has gained recognition in recent years. Unfortunately, only about one-third of known CRM1 cargo proteins are accessible in a single database since the last compilation in 2003. CRM1 cargo proteins are often recognized by a classical NES (leucine-rich NES), but this signal is notoriously difficult to predict from sequence alone. Fortunately, a recently developed prediction method, NESsential, is able to identify good candidates in some cases, enabling valuable hints to be gained by in silico prediction, but until now it has not been available through a web interface. We present ValidNESs, an integrated, up-to-date database holding 221 NES-containing proteins, combined with a web interface to prediction by NESsential.
INTRODUCTION
For many cellular and viral proteins, active transport is required for the journey from nucleus to cytoplasm through the nuclear pore complexes. This transport is mostly mediated by the karyopherin exportin 1/chromosomal region maintenance 1 (CRM1) recognizing the classical nuclear export signals (NESs) of cargo molecules. The classical NES is characterized by three to four conserved hydrophobic residues, usually leucine, and the spacing between them. Several consensus sequences have been proposed to describe the classical NES (1,2); however, as we previously demonstrated, they all suffer from poor predictive power in identifying potential NES-containing proteins (3). It should be noted that an increasing number of non-classical CRM1-mediated NESs, albeit still a minority, have been validated in recent years.
Many recent studies focus on the therapeutic potential of the CRM1-mediated nuclear export pathway. This nuclear export pathway is suggested to be involved in the mechanism inducing the abnormal localization of many tumor suppressors, p53 for instance, in various cancer cells (4). Furthermore, CRM1 has been found to be overexpressed in cervical cancer and critical for cancer cell proliferation and survival (5). As for the cargo proteins, many cellular NES-containing proteins are involved in important processes such as signal transduction, cell-cycle regulation and tumor suppression. Moreover, many known cargo proteins are viral, often playing a role in viral genome trafficking: the HIV-1 Rev protein is related to the export of unspliced or partially spliced viral messenger RNA (mRNA) (6); NS2/NEP of influenza A virus plays a critical role in the export of newly synthesized viral ribonucleoproteins, a complex composed of individual negative-sense viral RNAs and various viral proteins (7); while in adenovirus type 5, several NES-containing proteins were found to be required for efficient export of adenoviral early mRNA (8).
Due to their potential disease relevance, experimental identification of NES-containing proteins has been an active field of research. Surprisingly, this issue has been neglected by the computational biology community in recent years. NESbase (9), listing 75 validated NES-containing proteins has been a valuable resource for experimental and computational biologists, with >100 citations since its publication. Unfortunately, NESbase ceased updating after 2003 and now contains only about one-third of all validated NES-containing proteins. We therefore developed ValidNESs, in which we organize information on 221 NES-containing proteins compiled from the literature. Moreover, ValidNESs is easier to use and search against, is better cross-linked to external databases and provides a state-of-the-art prediction method in one site.
DATABASE CONTENT
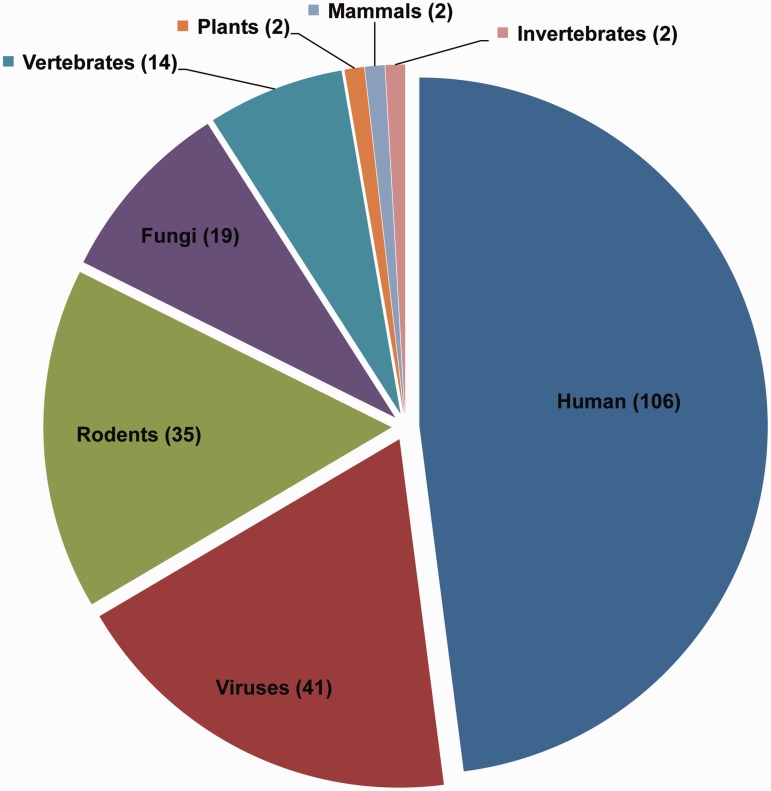
The first version of ValidNESs, made publicly available in June 2012, includes 262 functional NES sites from 221 NES-containing proteins (36 of them are multiple NES-containing proteins). In this version, we updated the collection of NES-containing proteins by compiling another 76 NES-containing proteins (up to 2012) and integrated them with those listed in NESbase (9) and the Supplementary Data of our previous NESsential paper (3), 75 and 70 proteins, respectively. Figure 1 shows a pie chart illustrating the number of proteins by species. In addition to sequence information, we collected a total of 52 local structures containing the entire NES region from the Protein Data Bank (PDB), which is exclusively available in ValidNESs. These local structures mainly (65%) consist of α-helix and other extended formations such as bends or loops. This result is basically consistent with the previous conclusion made from eight structures of NES-containing proteins (10). However, we found that β-structure can be found in 14 NES regions. Interestingly, Nilsen et al. (11) reported the first NES located on a β-strand in fibroblast growth factor-1 in 2007 and suggested that NESs with similar local structure should be found afterward. The updated data in ValidNESs support their speculation.
Figure 1.
Pie chart of species. Distribution of entries in ValidNESs. The number of species in which NES-containing proteins were validated are indicated in parenthesis.
To organize the data, we designed two different tables: one for NES-containing regions and another for NES-containing proteins. For users interested in functional NESs, sequence and secondary structural information (when applicable) can be found in the table of NES-containing regions. There is another table of NES-containing proteins designed for users requiring more information at the protein level, such as subcellular localization and protein–protein interaction. Detailed field descriptions for each table are given in Supplementary Tables S1 and S2, respectively.
THE CLASSICAL NES
Some previous work has defined a consensus sequence for NESs as [LIVFM]-x-(2,3)-[LIVFM]-x(2,3)-[LIVFM]-x-[LIVFM], where x is any amino acid (12). However, we found that 43% of NESs in ValidNESs deviate from this consensus sequence. We therefore defined a short consensus pattern [LIVFM]-x(2,3)-[LIVFM]-x-[LIVFM], hereafter denoted as the ‘NES motif’, containing the region bounded by the second and fourth hydrophobic positions of the former consensus (3), a region which has been shown to affect NES activity strongly (13,14). In ValidNESs, we use this generalized consensus pattern to divide experimentally determined NES sites into two categories: classical if the experimentally validated region contains or overlaps with a consensus match, otherwise non-classical. This definition of classical NES is justified by a dramatic improvement in sensitivity (from 57 to 86%). We tested the enrichment of this NES motif by binomial test, attaining P-values of 7.4e−64 (6-mer matches) and 1.5e−34 (7-mer matches), respectively. Finally, we generated sequence logos for the classical NESs aligned by consensus match (Figure 2).
Figure 2.
Sequence logos for NES sites. Sequence logos generated by the WebLogo server for NES motif matches after removing redundant sequences (with sequence identity >25%) and aligning the three hydrophobic positions within the motif. In general, the preference for negatively charged residues is lower than previously observed in NESbase. (A) Sequence logo for 6-mer NES motif matches with upstream and downstream 10-mer flanks (227 sites). (B) Sequence logo for 7-mer NES motif matches with upstream and downstream 10-mer flanks (162 sites).
DATA ACCESS
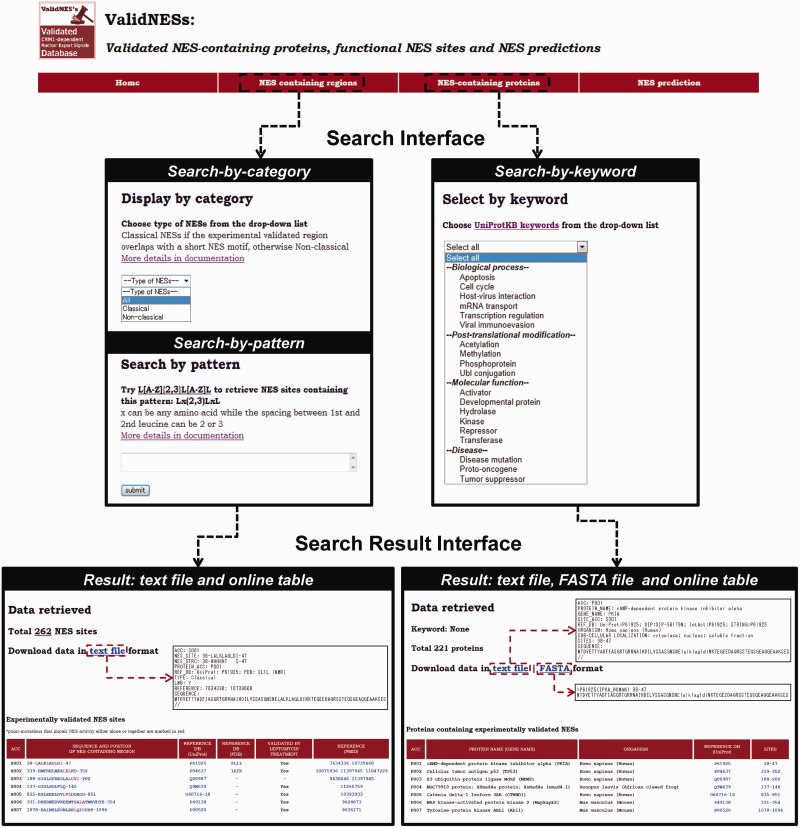
In addition to being up-to-date, ValidNESs provides an easy-to-use search interface. Table 1 summarizes the major difference between NESbase and ValidNESs. ValidNESs provides three search functions to retrieve particular data (or display all by default). Once the user submits the query, ValidNESs generates a complete table in text format ready for download and displays an online simplified table providing links to external databases. An overview of the search and search result interfaces is shown in Figure 3.
Table 1.
Comparison between NESbase and ValidNESs
| NESbase | ValidNESs | |
|---|---|---|
| Number of NES-containing proteins | 75 | 221a |
| Website architecture | HTML flat file | MySQL + PHP + Apache |
| Data access | No special search functionality | Searchable |
| User submission | Temporarily disabled | Supported |
aSeventy-five NES-containing proteins are imported from NESbase.
Figure 3.
An overview of the search and search result interfaces in ValidNESs. ValidNESs stores metadata in two tables and provides three search functions to access these data. Once users submit their queries, the search result in text file format and FASTA format (for table of NES-containing proteins only) is generated for download. Meanwhile, ValidNESs also displays an online table for quick browsing.
ValidNESs provides a ‘search-by-pattern’ function with regular expression support to facilitate retrieving particular NESs of interest. For example, Henderson and Eleftheriou (15) designed a Rev(1.4)-based shuttling assay and assessed the relative export efficiency of different types of NESs. This search function allows users to search and retrieve NES sites resembling those with available information on relative export efficiency. In ValidNESs, NES sites are divided into two categories based on the NES motif as previously mentioned. Therefore, users can use the ‘search-by-category’ function to retrieve the classical NES sites in an extended definition: that is, sites with an NES motif match lying inside or across the boundary of the experimentally determined NES-containing region. For NES-containing proteins, ValidNESs provides a ‘search-by-keyword’ function based on their UniProtKB keywords such as apoptosis or tumor suppressor. In addition to the complete table in text format, protein sequences including NES locations are also downloadable in FASTA format. Step-by-step instructions for novice users are available on the homepage of ValidNESs.
DATA CURATION
In most cases, the CRM1 dependence of NESs in ValidNESs is validated by treatment with leptomycin (LMB), a potent inhibitor blocking the binding of CRM1 to NESs (16). However, 42 (16%) of the NESs in ValidNESs have not had their CRM1 dependence validated with LMB. For these NESs, some other experimental techniques, such as yeast two-hybrid system and in vitro binding experiments, were used to demonstrate the interaction between CRM1- and NES-containing proteins (17,18). However, many of these NESs, 27 from NESbase for instance, were discovered around the early 2000s. In contrast, only 11 of these NESs were discovered in the last 5 years, as LMB has become widely used. For clarification, we add the LMB information in both the online and downloadable table of NES sites. We also cross-link to PDB in the same table if any structure containing the entire NES region is available. When multiple structures are available, we select the structure with the highest resolution and include the corresponding PDB ID in the table.
As mentioned above, 75 NES-containing proteins in ValidNESs were directly imported from NESbase. We updated the content in NESbase before integrating it into ValidNESs. This update includes one subsequently discovered NES for BRCA1 (19) and seven updated accession numbers in UniProtKB. In addition, we found nine protein sequences listed in NESbase differing from the current reference sequences in UniProtKB (eight with insertions and one with a point mutation). For these proteins, ValidNESs provides the sequences from UniProtKB and the modified NES positions according to the updated sequences. At the protein level, we provide information on subcellular localization and protein–protein interaction based on the relevant cross-references in UniProtKB. We extracted the GO cellular component annotation for the subcellular localization and imported the protein–protein interactions from four external databases: DIP (20), IntAct (21), MINT (22) and STRING (23). We also provide cross-references to NLSdb, a database of nuclear localization signals (NLSs) and nuclear proteins targeted to the nucleus by NLS motifs (24).
PREDICTION OF NES
ValidNESs provides online prediction of NES based on NESsential, our recently developed NES prediction method (3). Supplementary Figure S1 shows the submission interface where users can input a single protein sequence or a UniProt protein name (UniProt ID) such as IPKA_HUMAN. After successful submission and processing, users can view the prediction results, at both protein and site level, and an easy explanation about how to interpret them. ValidNESs currently allows one single sequence in a submission. For users having large computational needs such as large-scale screening, the standalone version of NESsential is recommended (http://seq.cbrc.jp/NESsential/).
DATA SUBMISSION
We greatly appreciate the efforts of researchers to discover and validate new CRM1-mediated NESs and encourage them to submit their new data to ValidNESs in the future. From the homepage of ValidNESs, we provide a preformatted form, including an example, for submission by email. We intend to maintain and frequently update ValidNESs for many years.
DISCUSSION
The large dataset consolidated in ValidNESs facilitates the investigation of various questions related to NES sequence and function. One interesting question is: why do some proteins have more than one NES? In 2007, Engelsma et al. (25) found a monomer-specific NES of human survivin, a key regulator of cell division containing two functional NESs, indicating that NESs in the same protein may play different functional roles. We therefore assume that distinct NESs in the same protein may be under different selective pressure to be conserved, e.g. some of them could be species specific. To test our assumption, we made an investigation among 28 multiple NES-containing proteins whose homologs are available in HomoloGene (http://www.ncbi.nlm.nih.gov/homologene). We defined an abrogation of an NES as a mutation which causes the NES to no longer match the NES motif covering the three essential hydrophobic residues. As a result, we found 13 out of 28 homologous groups containing at least one NES abrogation (see Supplementary Data), demonstrating that the presence of multiple functional NESs is not necessarily conserved in evolution.
CONCLUSION
We present ValidNESs, an integrated, up-to-date database and web interface to the NES prediction method NESsential. To illustrate the kind of analysis facilitated by the data organized in ValidNESs, we summarized the secondary structure propensity of NESs and discussed the existence of species-specific NESs. In conclusion, ValidNESs provides both updated data and an upgraded interface for convenient access to experimentally validated NESs- and NES-containing proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figure 1 and Supplementary Case Study.
FUNDING
National Science Council, Taiwan [NSC 99-2621-B-002-005-MY3 and 99-2621-B-010-001-MY3]; National Taiwan University Cutting-Edge Steering Research Project [10R70602C3 and 101R7602C3]; Top University Project [10R40044 and 101R4000]. Funding for open access charge: National Science Council, Taiwan [NSC 99-2621-B-002-005-MY3 and 99-2621-B-010-001-MY3]; National Taiwan University Cutting-Edge Steering Research Project [10R70602C3 and 101R7602C3].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are pleased to thank Dr Shunichi Kosugi for providing further supporting information of their original paper.
REFERENCES
- 1.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic. 2008;9:2053–2062. doi: 10.1111/j.1600-0854.2008.00825.x. [DOI] [PubMed] [Google Scholar]
- 3.Fu S-C, Imai K, Horton P. Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res. 2011;39:e111. doi: 10.1093/nar/gkr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner JG, Sullivan DM. CRM1-mediated nuclear export of proteins and drug resistance in cancer. Curr. Med. Chem. 2008;15:2648–2655. doi: 10.2174/092986708786242859. [DOI] [PubMed] [Google Scholar]
- 5.van der Watt PJ, Maske CP, Hendricks DT, Parker MI, Denny L, Govender D, Birrer MJ, Leaner VD. The Karyopherin proteins, Crm1 and Karyopherin β1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int. J. Cancer. 2009;124:1829–1840. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope TJ. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 1999;365:186–191. doi: 10.1006/abbi.1999.1207. [DOI] [PubMed] [Google Scholar]
- 7.Iwatsuki-Horimoto K, Horimoto T, Fujii Y, Kawaoka Y. Generation of influenza A Virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J. Virol. 2004;78:10149–10155. doi: 10.1128/JVI.78.18.10149-10155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid M, Gonzalez RA, Dobner T. CRM1-dependent transport supports cytoplasmic accumulation of adenoviral early transcripts. J. Virol. 2012;86:2282–2292. doi: 10.1128/JVI.06275-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 11.Nilsen T, Rosendal KR, Sørensen V, Wesche J, Olsnes S, Wiedłocha A. A nuclear export sequence located on a beta-strand in fibroblast growth factor-1. J. Biol. Chem. 2007;282:26245–26256. doi: 10.1074/jbc.M611234200. [DOI] [PubMed] [Google Scholar]
- 12.Kutay U, Güttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 14.Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- 15.Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 17.Neuber A, Franke J, Wittstruck A, Schlenstedt G, Sommer T, Stade K. Nuclear export receptor Xpo1/Crm1 is physically and functionally linked to the spindle pole body in budding yeast. Mol. Cell. Biol. 2008;28:5348–5358. doi: 10.1128/MCB.02043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson ME. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J. Biol. Chem. 2005;280:21854–21857. doi: 10.1074/jbc.M502676200. [DOI] [PubMed] [Google Scholar]
- 20.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair R, Carter P, Rost B. NLSdb: database of nuclear localization signals. Nucleic Acids Res. 2003;31:397–399. doi: 10.1093/nar/gkg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelsma D, Rodriguez JA, Fish A, Giaccone G, Fornerod M. Homodimerization antagonizes nuclear export of survivin. Traffic. 2007;8:1495–1502. doi: 10.1111/j.1600-0854.2007.00629.x. [DOI] [PubMed] [Google Scholar]