Abstract
The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) develops tools and resources that provide a structural view of biology for research and education. The RCSB PDB web site (http://www.rcsb.org) uses the curated 3D macromolecular data contained in the PDB archive to offer unique methods to access, report and visualize data. Recent activities have focused on improving methods for simple and complex searches of PDB data, creating specialized access to chemical component data and providing domain-based structural alignments. New educational resources are offered at the PDB-101 educational view of the main web site such as Author Profiles that display a researcher’s PDB entries in a timeline. To promote different kinds of access to the RCSB PDB, Web Services have been expanded, and an RCSB PDB Mobile application for the iPhone/iPad has been released. These improvements enable new opportunities for analyzing and understanding structure data.
INTRODUCTION
The RCSB Protein Data Bank (RCSB PDB) (1) provides access to the data in the PDB, the single archive of experimentally determined structures of nucleic acids, proteins and complex assemblies (2). The public archive currently contains >84 000 entries, derived data files and related data dictionaries. With >570 000 files, the PDB requires >130 GB of storage space. Data are updated weekly and loaded into the relational database that supports the web site.
The PDB is maintained by the members of the Worldwide PDB (wwPDB): RCSB PDB (USA) (1,3), PDB in Europe (PDBe, http://pdbe.org) (4), PDB Japan (PDBj, http://pdbj.org) (5) and BioMagResBank (http://bmrb.wisc.edu) (6). These member organizations host deposition, processing and distribution centers for PDB data. Data are deposited to the PDB, curated and annotated following wwPDB standards, and then made available on an FTP server. Each wwPDB partner offers unique ‘views’ of PDB data through the different query, analysis and visualization tools provided on their respective web sites.
The RCSB PDB web site currently hosts ∼240 000 unique visitors per month (based on the number of unique IP addresses), an increase from the 180 000 visitors last reported in 2011 (3). Web site users represent a variety of interests, including students (ranging from elementary school to graduate school), academic and industrial researchers, bench scientists and programers and web developers. To better serve these interests, the RCSB PDB home page and individual ‘Structure Summary’ pages can be customized by users by moving relevant data widgets (7) to different locations on the page, and hiding or minimizing areas of less interest. For education-focused browsing, a separate PDB-101 section offers related materials such as the ‘Molecule of the Month’ columns that tell the functional story of selected macromolecules.
PDB data can be searched in many different ways. The top menu bar can be used to perform simple searches, including author name, molecule name, sequence or ligand ID. ‘Advanced Search’ can be used to build queries with multiple constraints, such as ‘find all protein homodimers bound to DNA’. The ‘Browse Database’ option allows exploration of the PDB archive using different hierarchical trees. Browsers are available to search for related terms and structures based on many different classifications, such as Biological Process, Cellular Component, Molecular Function (8), Enzyme Commission number (http://www.chem.qmul.ac.uk/iubmb), Transporter Classification System (9), and structure classifications SCOP (10) and CATH (11). Data distribution summaries, shown as pie charts and lists of hyperlinks, are available for standard features of PDB entries (resolution, release date, experimental method, polymer type, organism and taxonomy). These drill-down distributions provide another way to browse and select data from the whole archive or any search results.
Query results can be refined, used to explore individual structures and exported to generate interactive and tabular reports. Tabular report features include online data sorting, column customization, filtering and output to other report formats. These reports also contain data from, and links to, external resources.
User feedback is an important influence on the evolution of the RCSB PDB resource. Recently added features, some developed based on this feedback, are described here.
NEW WEB SITE FEATURES
Simple searches
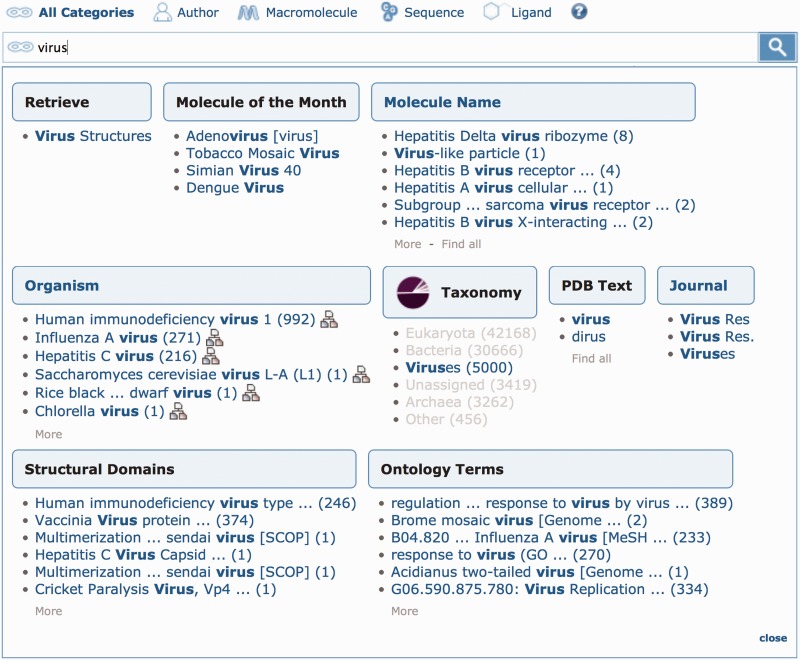
The most common uses of the web site are simple text searches. To further improve the text search, we have added an autocomplete feature to guide the user to more specific results. After typing a few letters in the top bar, a suggestion box organizes specific result sets in different categories. Each suggestion, which includes the number of results, links to the set of matching structures. Some of the suggestions use external data resources, such as the NCBI organism taxonomy tree (8,12). These possible matches can be especially helpful for finding structures when using common or vague search terms, as is shown in Figure 1 for the term ‘virus’.
Figure 1.
Top bar searching. This example shows several suggestions for the search term ‘virus’. In the Taxonomy category, the ‘Viruses’ link will return all entries in the virus superkingdom, even if the word ‘virus’ does not appear in the text of the entry. Conversely, entries with irrelevant matches for ‘virus’ (such as an occurrence in a related citation) are excluded. The searches with the most results are shown first, such as the hits for ‘Human immunodeficiency virus 1’ and ‘Influenza A virus’ under Organism. The ‘Molecule of the Month’ category offers related articles from PDB-101. Finally, a custom ‘Retrieve’ category provides easy access to all entries where the biological assembly represents the complete virus particle. Numbers in parentheses represent the number of entries that match a specific term, and text in brackets represents the name of a structural domain classification scheme or ontology. Search suggestions can be restricted to specific categories by selecting the ‘Author’, ‘Macromolecule’, ‘Sequence’ or ‘Ligand’ icon above the text search box. The default search is set to ‘All Categories’.
The top bar search is context-specific and intelligently detects the type of user input. Entering a sequence text string in the search box returns possible Basic Local Alignment Search Tool (13) search options. Chemical formulas and SMILES strings (14) are also recognized, e.g. the SMILES string for adenosine ‘Nc1ncnc2ncnc12’ yields choices of substructure, exact structure or structure similarity searches. If the suggestions are not what the user is looking for, it is still possible to perform a standard text search of the PDB entry (in mmCIF format) by pressing enter or clicking on the search icon.
Top bar simple searches can also be limited to specific categories by selecting the ‘Author’, ‘Macromolecule’, ‘Sequence’ or ‘Ligand’ icon. The ‘Author’ icon restricts searches to the names of depositors or primary citation authors. The ‘Macromolecule’ icon returns structures based on polymer names from the PDB and associated entries in cross-referenced sequence databases like UniProtKB (15). For example, typing ‘caspase’ provides suggestions for different types of caspases. By selecting ‘caspase-1’ and examining the PDB entries returned, it becomes obvious that the actual search is for PDB structures with cross-references to various UniProtKB entries for caspase-1 from different organisms. The ‘Sequence’ icon reveals a link to additional options for selecting the method and the parameters for a sequence search. Similarly, the ‘Ligand’ icon links to further options, including a chemical structure editor to draw a structure, and a form to search for ligands by name, identifier, formula and molecular weight.
New advanced search features
Advanced Search expands on the search functionality of the top bar searches by using additional and more specific data categories. Advanced Search has the capability of combining multiple searches of specific types of data in a logical AND or OR. The result is a list of structures that comply with ALL or ANY search criteria, respectively.
New Advanced Search options are available to search by: ‘All/Experimental Type/Molecule Type’ to quickly access all PDB entries or a subset based on experimental and macromolecular type, structure determination/phasing method (e.g. molecular replacement, MAD or SAD), ‘Link Records’ to find structures containing inter-residue connectivity (LINK records in PDB entries) that cannot be inferred from the primary structure, structures determined by electron microscopy for which experimental data files are available in the PDB or at the Electron Microscopy DataBank (16) and Pfam ID (17).
All Advanced Search query results can be further refined, filtered to remove similar sequences or used to generate reports.
Structure alignments
Sequence and structure alignments are standard methods for analyzing the evolutionary and functional relationship between proteins (18–23). The Protein Comparison Tool offers a number of sequence and structure alignment algorithms for a detailed analysis of pairwise relationships (24). Additional algorithms are available via submission of alignments to some of the leading external web servers (25–28). The Protein Comparison Tool has also been used to provide the pre-calculated alignments, updated weekly, of a representative subset (based on sequence identity) of the PDB (24). The first version of this tool was based on alignments of whole protein chains. This has recently been refined to provide alignments on a domain basis.
The calculation based on domains extends our sequence clustering approach. To remove redundancy, we start with a 40% sequence identity clustering procedure based on complete polypeptide chains, and select a representative chain from each sequence cluster (3). If the representative chain contains multiple domains, each is included. SCOP 1.75 domain assignments are used when available; otherwise, assignments are computed using ProteinDomainParser (PDP) (29). Pairwise alignments of the domains are performed with the jFatCat version (24) of FatCat (22).
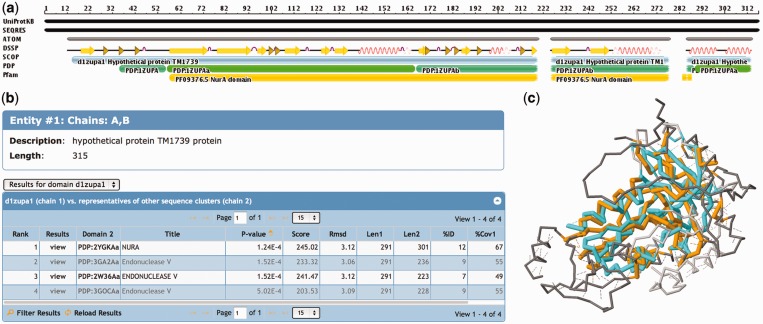
For each PDB entry, the ‘3D Similarity’ tab provides a visual summary of the protein chains. Figure 2 highlights how the residues listed in the sequence (SEQRES) and in the atom records (ATOM) map onto the relevant parts of the UniProtKB sequence, along with annotations from DSSP (32), SCOP, PDP (29) and Pfam (33).
Figure 2.
Domain-based structural alignment database search results for PDB ID 1ZUP (30), a hypothetical protein and a putative nuclease. (a) The sequence is shown with different annotations, starting with the UniProtKB sequence at the top. Most of this sequence has been resolved (SEQRES) with ATOM records available in the PDB, with the exception of a region at the N-terminus and two short regions toward the C-terminus of the protein. SCOP provides an annotation of a single domain protein, the PDP software assigns two domains to this protein and Pfam detects a NurA domain motif. (b) The hits listed in the table show similarities to a NurA structure, as well as several endonucleases. (c) When viewing the details of the first alignment [1ZUP shown in orange and NurA domain of 2YGK (31) in cyan], one notices that several loop and helical regions are not conserved between the two structures, but the core is well conserved. Structural alignments are accessible from the 3D Similarity tab of any entry’s Structure Summary page.
The results of the pre-calculated database searches are shown in a table that displays the most important calculated alignment scores (Figure 2). For multi-domain proteins, it is possible to switch between the results for different domains by selecting a domain from the pull-down menu above the table, or by clicking on a domain in the sequence image.
The results table can be sorted and filtered, and links to the 3D structure alignment in Jmol (http://www.jmol.org) (34) (Figure 2) and to information about similar domains.
Ligand reporting and visualization
Information about the chemistry and structure of all small molecule components found the PDB is contained in the Chemical Component Dictionary maintained by the wwPDB at wwpdb.org (35). As described earlier, specialized ligand queries can be made using the top bar search or Advanced Search. Special support is also offered for the analysis of ligands associated with PDB entries. The RCSB PDB web site builds on the functionality developed for the small molecule resource Ligand Expo (http://ligand-expo.rcsb.org) (36) by providing special support for the analysis of ligands associated with PDB entries.
Any ligands included with a PDB entry are listed in the ‘Ligand Chemical Component’ widget of the entry’s ‘Structure Summary’ page. This area displays the name and formula of each ligand, links to the summary page for the ligand and provides access to 3D visualization of the ligand in the context of that particular PDB entry using the Ligand Explorer viewer (37). For non-trivial ligands, a PoseView (38) interaction diagram shows which atoms or areas of the ligand and the polymer interact with each other, as well as the type of interaction.
‘Ligand Summary’ pages are organized into widgets highlighting different types of hyperlinked information, similar to Structure Summary pages for individual PDB entries. These widgets provide an overview of the ligand, with links to PDB entries where the component appears as a non-polymer or as a non-standard component of a polymer, links to ligand summary pages for similar ligands and stereoisomers, 2D and 3D visualization and links to many external resources. Ligand Summary pages also display information about molecules that have been annotated as having sub-components. For example, the summary page for ligand 0GM lists the sub-components with identifiers BNA, GLU, STA, LEU and TRJ that are connected with peptide-like or other bonds.
Ligand Summary Reports can be generated for query result sets and downloaded in a text file or a spreadsheet. These reports include information about the selected ligands, such as formula, molecular weight, name, SMILES string, which PDB entries are related to the ligand and how they are related. Each ligand included in the report can be expanded to show a sub-table of all related PDB entries that contain the ligand, the entries that contain the ligand as a free ligand and entries that contain the ligand as part of a polymer.
Visualization of molecular surfaces
Protein Workshop (37) is one of several 3D molecular viewers offered from the RCSB PDB web site. It offers quick default styles and views, with additional appearance options. Chains and atoms can be selected by either clicking on the structure or molecules displayed as a tree.
Protein Workshop now supports molecular surfaces to aid in the display of quaternary structure, protein–protein interactions and binding sites. Surfaces are created for all macromolecule chains in a PDB entry using the Euclidean distance transform algorithm from Xu and Zhang (39). For biological assemblies, surfaces are generated using the symmetry operation of the space group, which allows the display of even the largest assemblies in the PDB [i.e. the PBCV-1 virus capsid with 5040 chains, PDB ID 1M4X (40)] on a standard laptop computer. Surfaces can be color coded by chain, entity (unique macromolecules) and hydrophobicity. Color-blind friendly color schemes were adopted from ColorBrewer, a tool for selecting color schemes for maps (41). In addition, options to export high-resolution images with custom sizes for publications and posters are available for the three RCSB PDB viewers: Protein Workshop, Simple Viewer and Ligand Explorer.
WEB SERVICES
Web Services are used by software tools that efficiently and remotely interact with PDB data on the fly, eliminating the need for local data storage. The RCSB PDB hosts RESTful search and fetch services that return XML files in response to URL requests. Search services return PDB ID lists for queries based on Advanced Search queries. Fetch services return data (such as entity descriptions, ligand information and external annotations) for a given list of IDs. In addition to the services reported previously (3), new services are described in Table 1. For example, access to sequences released ahead of the structure is now frequently used by structure prediction servers for blind predictions (such as http://www.cameo3d.org/). More than 100 data fields can be exported in a generic way using the tabular report service. For example the URL
specifies a Web Service request for a list of PDB IDs with four data fields in the comma-separated value file format.
Table 1.
Recently introduced RESTful Web Services
| Web service | Description |
|---|---|
| Pre-released sequences | Access sequences in FASTA format for entries that have been deposited to the PDB, but are on hold until publication or a specified release date. |
| Custom reports | Create tables of sequence, structure, function, ligand information, experimental details and structure annotations in comma-separated value file, XML or MSExcel format. |
| Pfam annotations | Retrieve Pfam domain annotations, calculated by running Pfam’s Hidden Markov Models (42). |
| Domain-based structural alignments | Retrieve structural neighbors and alignment scores. |
A full list of web services and examples are available at: http://www.rcsb.org/pdb/software/rest.do.
RCSB PDB MOBILE
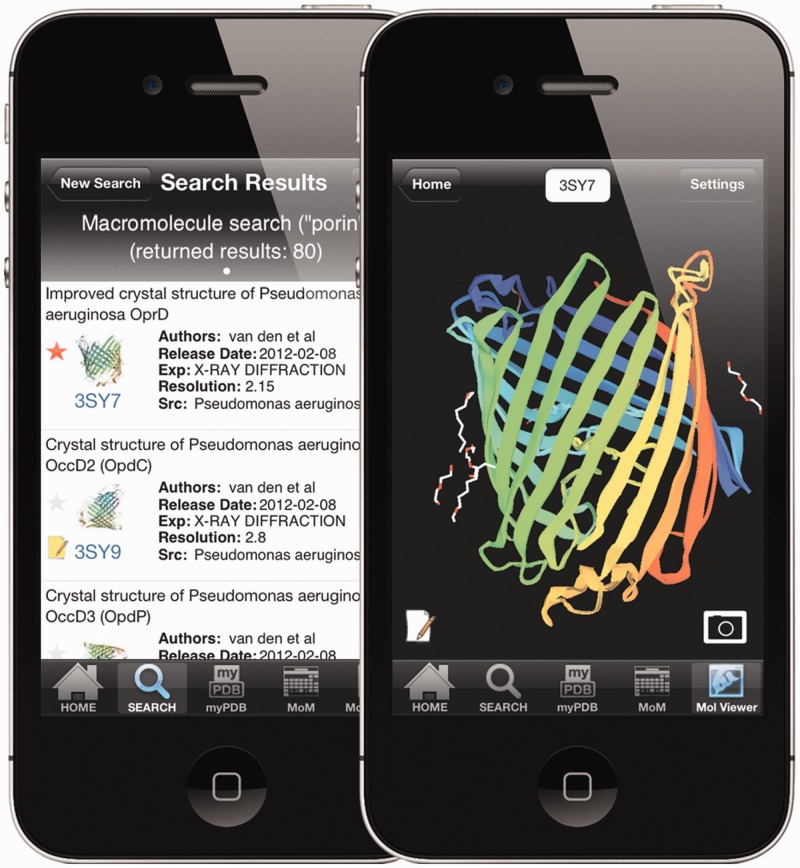
A simplified interface to the RCSB PDB is available as an app for the iPhone/iPod and the iPad (Figure 3). The app offers special features, including a simplified search for macromolecule name, author name and PDB ID. Query results, displayed in a single page listing, can be filtered by author name, title and organism. A macromolecule image and the PubMed abstract (when available) for individual entries are displayed when the user selects an entry from a returned query results list.
Figure 3.
RCSB PDB Mobile. The left image shows the query results from a macromolecule name search for ‘porin’ on an iPhone. Basic information about each entry is displayed in the results list. Entry 3SY7 has been tagged (red asterisk) and entry 3SY9 (43) has been annotated (yellow note pad icon) in the MyPDB account of the user. Entry 3SY7 in the NDKMol viewer is shown on the right. Structures in the viewer can be rotated, translated and zoomed using finger gestures, and touching the camera icon captures an image. The menu at the bottom allows the user to switch to the search menu, run MyPDB queries, browse ‘Molecule of the Month’ articles or launch the 3D viewer.
RCSB PDB Mobile also provides a listing of the most recently released PDB entries, and can be used to explore the archive of ‘Molecule of the Month’ articles and RCSB PDB news. Users can connect to their MyPDB account, a service that allows users to store queries and structure annotations.
RCSB PDB Mobile includes an integrated molecular viewer, NDKMol, developed by collaborator Dr. Takanori Nakane, Kyoto University. The viewer presents an interactive molecular rendering using downloaded PDB format files. The user is able to modify the appearance of the rendering by changing display settings such as display style (Ribbon, C-alpha trace, strand or B-factor tube), ligand/HET atom style (sphere, stick or line), nucleotide base style (line or polygon), color scheme (spectrum, by chain by secondary structure, polar/non-polar or B-factor), symmetry mates (biological assembly or crystal packing) and several other options.
A version of the app for the Android platform is in development.
PDB-101: EDUCATIONAL FEATURES
The volume and complexity of PDB data can pose a challenge for users, particularly beginning students. To support non-experts interested in exploring biomolecular structure, RCSB PDB educational resources and features (44,45) have been packaged together to form the ‘PDB-101’ web site that is accessible from the main web site via the PDB-101 logo. PDB-101 currently supports five main features: the archive of ‘Molecule of the Month’ columns, which describe biomolecular structure and function for general audiences; Educational Resources, including posters and animations; the ‘Understanding PDB Data’ resource for learning about data files and structure determination methods; the Structural View of Biology browser and Author Profiles.
Structural view of biology
The Structural View of Biology, shown at the PDB-101 landing page, was designed to encourage self-guided exploration of the PDB by non-experts. It is separated into six functional categories, such as ‘Enzymes’ and ‘Protein Synthesis’, and allows users to browse based on the biological properties typically used in biology and chemistry education. The topics can be browsed down to individual ‘Molecule of the Month’ features, which include annotated Jmol views and links to simplified summary pages highlighting specific example entries. This provides novice users with a subset of the PDB archive selected for its utility in education.
Author profiles
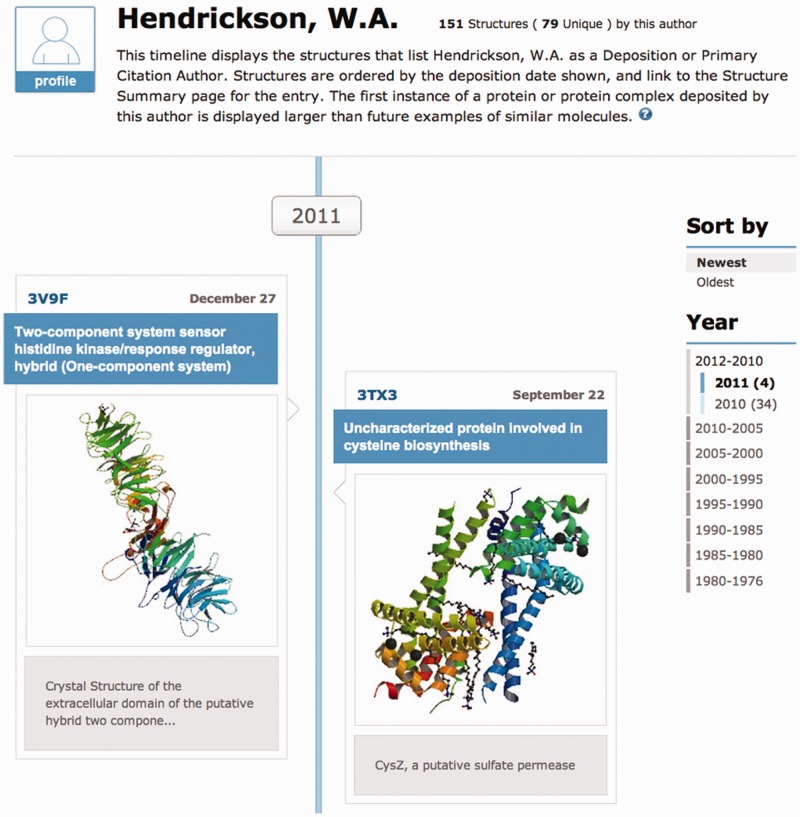
A unique historical and educational tool enabled by the database, ‘Author Profile’ displays a vertical timeline of the structures associated with either an individual author or a structural genomics center (Figure 4). A text search form is available to find different profiles. The structures shown are selected based on author name (deposition or primary citation author), and ordered by deposition date. Unique structures, denoted by a blue background and shown with a large image, indicate the first structure of a polymer or polymer complex deposited by the researcher. Subsequent structures that contain the same set of UniProtKB cross-reference identifiers (15) are displayed with a smaller image.
Figure 4.
The top portion of an Author Profile displaying the structures associated with author W.A. Hendrickson is shown. Timelines can be sorted by deposition date and specific time ranges can be selected from the right hand menu. Author profiles can be bookmarked and shared.
SUMMARY
We continue to build and improve RCSB PDB resources to enable a structural view of biology. New search options include search suggestions and Advanced Search options that guide the user to more specific search results. The Author Profile tool offers a new way to explore structures solved by individual authors and structural genomics centers. Structural alignments are now available for representative domains, rather than just protein chains. Ligand searching, reporting, and visualization has been improved. The addition of surfaces to the 3D viewers enables the analysis of ligand binding sites, protein–protein interactions and quaternary structure. Web Services have been expanded to include pre-release sequences and a generic mechanism to retrieve PDB data through tabular report services. To cater to the rapidly growing number of mobile users, we have deployed RCSB PDB Mobile for the iPhone and iPad, and an Android version is under development. A new educational section, PDB-101, hosts the educational content and provides a hierarchy to browse ‘Molecule of the Month’ articles. New web site releases are announced on the ‘What’s New’ widget on the home page, and in weekly news announcements.
FUNDING
National Science Foundation [NSF DBI 0829586]; National Institute of General Medical Sciences (NIGMS); Office of Science, Department of Energy (DOE); National Library of Medicine (NLM); National Cancer Institute (NCI); National Institute of Neurological Disorders and Stroke (NINDS); National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Computational resources for structural alignments are provided in part by the Open Science Grid (http://www.opensciencegrid.org) funded by the National Science Foundation; and the Office of Science, Department of Energy (DOE) [NSF 0753335]. Funding for open access charge: National Science Foundation [NSF DBI 0829586].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank BioSolveIT GmbH (http://www.biosolveit.de) for access to PoseView, and ChemAxon (http://www.chemaxon.com) for providing Marvin Sketch, JChem Base and Standardizer for the chemical structure search. Dong Xu and Yang Zhang provided source code for the Euclidean distance transform algorithm for calculating molecular surfaces. Takanori Nakane developed an Objective-C version of the NDKViewer for the RCSB PDB Mobile. Access to binding affinity data was provided by Michael Gilson (BindingDB), Heather Carlson (BindingMOAD) and Renxiao Wang (PDBbind-CN). In addition, we also thank all users who provided feedback, and RCSB PDB staff, past and present, for suggestions, critical review and testing of new features. The RCSB PDB is managed by two members of the RCSB: Rutgers and UCSD, and is a member of the wwPDB.
REFERENCES
- 1.Berman HM, Westbrook JD, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 3.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Golovin A, Gore SP, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2012;40:D445–D452. doi: 10.1093/nar/gkr998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinjo AR, Suzuki H, Yamashita R, Ikegawa Y, Kudou T, Igarashi R, Kengaku Y, Cho H, Standley DM, Nakagawa A, et al. Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res. 2012;40:D453–D460. doi: 10.1093/nar/gkr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne PE, Beran B, Bi C, Bluhm W, Dunbrack R, Prlic A, Quinn G, Rose P, Shah R, Tao W, et al. Will widgets and semantic tagging change computational biology? PLoS Comput. Biol. 2010;6:e1000673. doi: 10.1371/journal.pcbi.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Gene Ontology Consortium. The gene ontology: enhancements for 2011. Nucleic Acids Res. 2012;40:D559–D564. doi: 10.1093/nar/gkr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The transporter classification database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 11.Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weininger D. SMILES 1. Introduction and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31. [Google Scholar]
- 15.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 20.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Tatusova TA, Madden TL. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 22.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19:ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 23.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatory extension of the optimum path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 24.Prlic A, Bliven S, Rose PW, Bluhm WF, Bizon C, Godzik A, Bourne PE. Pre-calculated protein structure alignments at the RCSB PDB website. Bioinformatics. 2010;26:2983–2985. doi: 10.1093/bioinformatics/btq572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godzik A. Fold recognition methods. Methods Biochem. Anal. 2003;44:525–546. doi: 10.1002/0471721204.ch26. [DOI] [PubMed] [Google Scholar]
- 26.Park BJ, Park JI, Byun DS, Park JH, Chi SG. Mitogenic conversion of transforming growth factor-beta1 effect by oncogenic Ha-Ras-induced activation of the mitogen-activated protein kinase signaling pathway in human prostate cancer. Cancer Res. 2000;60:3031–3038. [PubMed] [Google Scholar]
- 27.Sippl MJ, Wiederstein M. Detection of spatial correlations in protein structures and molecular complexes. Structure. 2012;20:718–728. doi: 10.1016/j.str.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrov N, Shindyalov I. PDP: protein domain parser. Bioinformatics. 2003;19:429–430. doi: 10.1093/bioinformatics/btg006. [DOI] [PubMed] [Google Scholar]
- 30.Joint Center for Structural Genomics. Crystal structure of hypothetical protein (tm1739) from Thermotoga maritima at 2.20 Å resolution. Proteins. 2005;61:669–673. doi: 10.1002/prot.20542. [DOI] [PubMed] [Google Scholar]
- 31.Blackwood JK, Rzechorzek NJ, Abrams AS, Maman JD, Pellegrini L, Robinson NP. Structural and functional insights into DNA-end processing by the archaeal HerA helicase-NurA nuclease complex. Nucleic Acids Res. 2012;40:3183–3196. doi: 10.1093/nar/gkr1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 33.Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson RM. Jmol—a paradigm shift in crystallographic visualization. J. Appl. Cryst. 2010;43:1250–1260. [Google Scholar]
- 35.Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, et al. Remediation of the Protein Data Bank Archive. Nucleic Acids Res. 2008;36:D426–D433. doi: 10.1093/nar/gkm937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Z, Chen L, Maddula H, Akcan O, Oughtred R, Berman HM, Westbrook J. Ligand depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20:2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 37.Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stierand K, Rarey M. Drawing the PDB: protein−ligand complexes in two dimensions. Med. Chem. Lett. 2010;1:540–545. doi: 10.1021/ml100164p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu D, Zhang Y. Generating triangulated macromolecular surfaces by Euclidean Distance Transform. PLoS One. 2009;4:e8140. doi: 10.1371/journal.pone.0008140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, Rossmann MG. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl Acad. Sci. USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrower M, Brewer CA. ColorBrewer.org: an online tool for selecting colour schemes for maps. Cartogr. J. 2003;40:27–37. [Google Scholar]
- 42.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eren E, Vijayaraghavan J, Liu J, Cheneke BR, Touw DS, Lepore BW, Indic M, Movileanu L, van den Berg B. Substrate specificity within a family of outer membrane carboxylate channels. PLoS Biol. 2012;10:e1001242. doi: 10.1371/journal.pbio.1001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dutta S, Zardecki C, Goodsell D, Berman HM. Promoting a structural view of biology for varied audiences: an overview of RCSB PDB resources and experiences. J. Appl. Cryst. 2010;43:1224–1229. doi: 10.1107/S002188981002371X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zardecki C. Interesting structures: education and outreach at the RCSB Protein Data Bank. PLoS Biol. 2008;6:e117. doi: 10.1371/journal.pbio.0060117. [DOI] [PMC free article] [PubMed] [Google Scholar]