Abstract
The National Center for Biotechnology Information (NCBI) Clone DB (http://www.ncbi.nlm.nih.gov/clone/) is an integrated resource providing information about and facilitating access to clones, which serve as valuable research reagents in many fields, including genome sequencing and variation analysis. Clone DB represents an expansion and replacement of the former NCBI Clone Registry and has records for genomic and cell-based libraries and clones representing more than 100 different eukaryotic taxa. Records provide details of library construction, associated sequences, map positions and information about resource distribution. Clone DB is indexed in the NCBI Entrez system and can be queried by fields that include organism, clone name, gene name and sequence identifier. Whenever possible, genomic clones are mapped to reference assemblies and their map positions provided in clone records. Clones mapping to specific genomic regions can also be searched for using the NCBI Clone Finder tool, which accepts queries based on sequence coordinates or features such as gene or transcript names. Clone DB makes reports of library, clone and placement data on its FTP site available for download. With Clone DB, users now have available to them a centralized resource that provides them with the tools they will need to make use of these important research reagents.
INTRODUCTION
The availability of genome sequences for many organisms has transformed the way researchers can approach biological questions. However, there remains a need to associate such genome sequences with physical reagents that can be used to perform experiments. Clone DB, the former Clone Registry database hosted by the National Center for Biotechnology Information (NCBI), recently underwent a series of major updates to improve its role as a resource for one important such reagent, namely clones (1). Clone DB now provides integrated information for both vector-based genomic and cell-based clones, including sequence data, map positions, gene content and distributor information (http://www.ncbi.nlm.nih.gov/clone/).
NCBI established the original Clone Registry database during the era of the human and mouse sequencing projects to help genome centers manage sequencing of clones in assembly tiling paths. Notably, the Clone Registry also established a standardized naming scheme for genomic clones that was adopted by many of the sequencing centers and facilitated the consolidation of various clone-associated data found in different NCBI databases within the Clone Registry. Early human and mouse clone records included information about associated insert and end sequences, genetic markers present on clones and mapping information. Additionally, they provided contact information for clone distributors. However, the database only represented genomic clones, and while it later expanded to include clone data from additional eukaryotic organisms, records for most of these additional species lacked the depth found in the human and mouse clone records. It was therefore clear that database updates were needed in order for the Clone Registry to remain a useful resource to the research community at large, especially once the initial human and mouse genome sequencing projects were completed.
Even in this era of next-generation sequencing technologies, the need for a resource such as Clone DB persists, as clones continue to play a vital role in biological research. In species where whole-genome sequencing has largely replaced clone-based tiling paths as the basis for genome assembly, the alignment of genomic clone end sequences to these assemblies remains an important mechanism for assessing assembly quality. Clone-based sequences are also commonly used in conjunction with whole-genome sequencing in complex genomic regions (2–5). In addition, end sequence profiling is a valuable technique for the discovery and analysis of structural variation (6,7). Notably, genomic clones remain the sequencing reagent of choice for many organisms in which large amounts of variation or repetitive content largely confound whole-genome assembly [The Danio rerio Sequencing Project (http://www.sanger.ac.uk/Projects/D_rerio/), (8)]. Cell-based reagents, such as gene trap and gene-targeting clones, also continue to serve as valuable biological resources. In both animal and plant species, libraries of such clones are routinely used to investigate genotype–phenotype relationships and define genetic pathways (9–12). Thus, the need for Clone DB, a clone-centric database that integrates data associated with various clone types, remains as important as ever. NCBI, through its updated Clone DB, now provides a resource that consolidates much of the available clone-related sequencing data found in the primary data archives, layers on mapping and gene information where possible and provides library construction and clone distribution details so that researchers can acquire the clone reagents they need for their research. The clone-centric Clone DB will facilitate the use of these valuable research reagents.
DATABASE DESCRIPTION
Database design
Clone DB is a relational database implemented in MSSQL. Although Clone DB retains many of the features and functionality of the Clone Registry, its databases and tables were entirely redesigned to accommodate the broader range of clone types and metadata represented in Clone DB. Clone and library records, along with their associated metadata are stored within a single database. Clone placements, as well as the assembly metadata needed to generate such placements are maintained in separate databases on the same server. Unique library and clone ids are shared by all of these databases. Library and clone records from the Clone Registry database were copied to Clone DB for its initial population. Clone and library records in Clone DB are indexed into NCBI’s Entrez search and retrieval system.
Data types and organization
Libraries
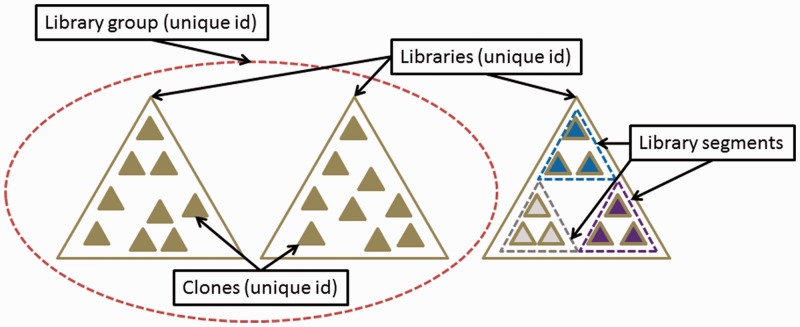
Operationally, libraries are collections of clones, and Clone DB utilizes this hierarchical relationship to organize data. All clone records, regardless of clone type, must be associated with a library record (Figure 1). Within the database, a library record is uniquely defined by the combination of the library name and species. Each library record is assigned a unique numeric identifier. For genomic libraries, metadata collected includes details about source DNA, library construction, library statistics, alternate names and abbreviations by which the library is known and library distributors, as well as publications describing the construction or end sequencing of the library. Metadata captured for cell-based libraries includes the library creator, cloning vector, parental cell line, parental cell line strain and allele type. Libraries sharing one or more user-defined features can be assigned to a library group. This has proven useful for making associations between libraries created by various groups as part of larger collaborations, such as those from the International Knockout Mouse Consortium (IKMC) or International Gene Trap Consortium (IGTC). Within a library record, library segments are used to distinguish subsets of clones that share common attributes within the larger context of the library. For example, the so-called ‘CHORI-105 Trypanosoma cruzi BAC Library’ (http://www.ncbi.nlm.nih.gov/clone/library/genomic/145/), actually contains both BAC and fosmid clones. The definition of two library segments for this record permits the database to maintain library construction details specific to each of the two vector types represented by the library.
Figure 1.
Schematic illustrating data relationships in Clone DB. Three different libraries (large open triangles) are represented. Libraries sharing a common set of user-specified attributes can be assigned to a library group (dashed circle). Within Clone DB, every clone (small filled triangles) must be associated with an existing library record. In instances where subsets of clones belonging to the same library exhibit different attributes (small triangles at far right with different fill colors), library segments are used to distinguish clones sharing common attributes (dashed open triangles). Library groups, libraries and clones are all assigned unique numeric identifiers within Clone DB; library segments are attributes of library records.
A primary goal of Clone DB is to enable researchers to make connections between data and physical reagents. Therefore, the libraries for which records are maintained in Clone DB predominantly represent those available from commercial or academic distributors (including individual investigators), along with a limited collection of libraries deemed to have sufficient scientific significance to warrant representation even in the absence of their distribution (e.g. they contribute sequence to a reference assembly or have many publicly available sequences). The current collection of 327 murine cell-based library records in Clone DB, along with their associated metadata, is provided by Mouse Genome Informatics [MGI (http://www.informatics.jax.org/)] and is updated on a weekly basis. These records represent all of the gene trap and gene target libraries produced by the IKMC (9,13,14) and IGTC (15), as well as the Lexicon Genetics collection of gene traps (16). Genomic library records in Clone DB include the original set of libraries imported from the Clone Registry database, as well as new library records generated by database curators. New records represent many of the libraries available from the larger domestic and international commercial and academic distributors, whom curators actively solicit for genomic library information. Additional curatorial emphasis is placed on providing representation for genomic libraries that contribute to the generation of reference assemblies, have been extensively end or insert sequenced and those that have been fingerprinted. Curators continue to update the database with new library records, placing priority on those whose representation is specifically requested by users contacting the Clone DB (clonereg-admin@ncbi.nlm.nih.gov). At the time of writing, Clone DB has records for 540 genomic libraries, representing nearly 150 different organisms, of which more than half are plants.
Clones
Although libraries serve as a data object used for organization in Clone DB, it is the clones belonging to these libraries that comprise the other data object in this database. All clone records receive a unique database identifier and must be associated with a library record (Figure 1). The mere physical existence of a clone is not sufficient to warrant the creation of a clone record in the database. Genomic clone records are only created for the subset of clones in a library for which sequence or mapping data is available. A major feature of the clone record is the clone name, which is the attribute by which most researchers and distributors identify these resources. The assignment of names to genomic clones poses a significant challenge, as it is not uncommon to find that different submitters have provided different permutations of a clone name on different data submissions that represent the same clone object. Whenever possible, Clone DB attempts to parse submitter-provided names and assign a standardized name comprised of the clone’s microtiter plate address (plate number, row and column), prefixed by the Clone DB library abbreviation to each record. The submitter-provided name then is stored as an alias of the standard name. If such a standard name cannot be parsed from the submitter-provided name, the submitter-provided name is assigned as the clone name. All names and aliases associated with genomic clone records are indexed and can be used as search queries. Additional information about clone name and data management in Clone DB is provided in via the website (http://www.ncbi.nlm.nih.gov/clone/content/overview/).
Data associated with genomic clone records in Clone DB are derived from a variety of sources. To identify insert sequences associated with genomic clones, Clone DB queries the INSDC nucleotide databases to retrieve high-throughput genomic sequences and their associated metadata for all organisms that have at least one library represented in the database. End sequences and their metadata are retrieved from both the dbGSS and Trace Archive databases (1), using library-specific queries. Insert and end sequence records containing clone names that cannot be associated with existing clone records, or from which new standardized clone names cannot be parsed and created, are flagged for curatorial review. Clone DB provides database cross-references that expose these clone-to-sequence relationships. Fingerprint data for genomic clones is externally derived, coming from the FPC database maintained by the Michael Smith Genome Sciences Centre in Vancouver, Canada [http://www.bcgsc.ca/data/data, (17)]. Likewise, data for cytogenetic map positions and sequence-tagged sites (STS) markers mapped to human genomic clones are taken from the work of the BAC resource consortium [http://www.ncbi.nlm.nih.gov/genome/cyto/hbrc.shtml, (18)] and National Cancer Institute’s Cancer Chromosome Aberration Project [http://cgap.nci.nih.gov/Chromosomes/CCAP_BAC_Clones, (19)].
Clone DB obtains the set of murine cell-based clones for which records will be provided from weekly reports provided by MGI and adopts the MGI-provided clone names. MGI also contributes clone library, gene, allele and sequence-identifier information, as well as creator and distributor details for all murine cell-based clone records (20,21) in Clone DB.
Genomic clone placements
The collection of clones in a genomic library represents an unassembled set of DNA that comprises an organism’s genome. In general, a clone’s utility as a research reagent is dependent upon whether it can be placed in genomic context and in the context of other clones. Doing so enables a researcher to make certain assumptions about the sequence and gene content represented in the clone and about the genome from which it is derived. Thus, in order to add value to the clones represented in the database, Clone DB maps genomic clones to RefSeq assemblies (22) that have been annotated via the NCBI annotation pipeline. Clones are generally mapped on a target genome that is the same species as the library DNA source, but cross-species placements may also be generated. For example, sheep genomic clones have been mapped to various cow assemblies, as the cow represents the nearest species for which an assembled RefSeq genome is available.
Currently, all placements generated by Clone DB are based entirely on end sequence alignments. End sequences associated with clone records are screened to remove vector contamination and low-quality bases. The set of cleaned ends is aligned to a Window-Masked (23) genome assembly of interest using an in-house, BLAST-derived alignment algorithm called NG Aligner. Clone placements are subsequently generated by the pairing of end placements representing the forward (F) and reverse (R) ends of a given clone. The current NCBI clone placement algorithm utilizes two approaches to minimize self-overlapping clone placements and present users with the most likely placement(s) for each clone. First, in instances where there are overlapping end placements for sequences associated with a given clone end, NCBI clusters the overlapping end placements and selects a single prototype for use in clone placements. The prototype is the end placement that holds the 5′-outermost position on the scaffold to which it aligns. No other end placements in the cluster contribute to clone placements. Second, in instances when prototypes contribute to a set of self-overlapping clone placements, NCBI selects a single clone placement from the set as an archetypal placement. Among clones with concordant clone placements, the archetype represents the concordant clone placement comprised of the best ranked pair of end placements within that set. If no concordant placement exists, the archetype represents the discordant clone placement comprised of the best-ranked pair of end placements. A clone may have more than one archetypal placement, but they may not overlap. Only archetypal placements are reported and displayed in clone records. To define the average insert size and standard deviation for each clone library, Clone DB uses only the subset of clone placements in which: (i) the placement is comprised of one forward and one reverse end, (ii) both ends are uniquely placed, (iii) both ends are placed on the same assembly scaffold, (iv) the end placements face each other and (v) the placement length is between 50 and 500 kb (BAC/PAC) or 10 and 100 kb (fosmids). Clone placement concordance is determined on the basis of placement length (within 3 SD of the library average) and the orientation of the contributing end placements (facing one another on opposite strands). In addition, clone placements are assigned a score that reflects the confidence level of the placement. This score is determined by the number of placements for the clone and whether there are additional end placements that do not support the clone placement. More information about the clone placement algorithm can be found in documentation on the Clone DB website (http://www.ncbi.nlm.nih.gov/clone/content/placements/). As described below, clone placement results are provided in both tabular and graphic formats in the displays for individual clone records, and as well as in the NCBI Clone Finder tool.
Accessing data
Searching Clone DB
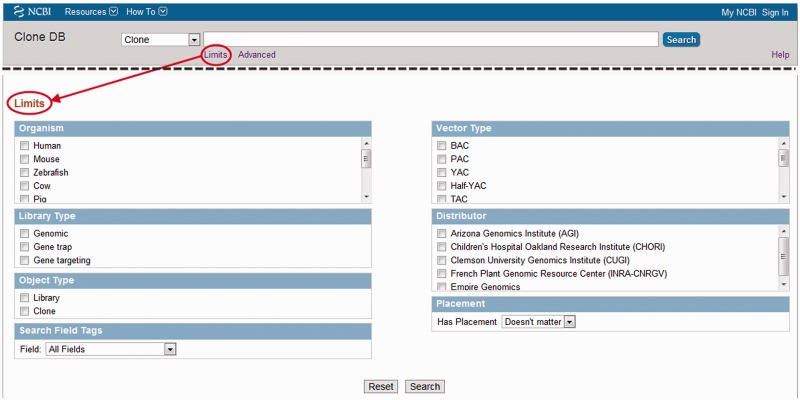
Clone DB is an NCBI Entrez database and queries can be initiated by entering free text into the search box located at the top of each webpage (Figure 2). Clone DB has been indexed to allow users to compose complex search queries using a wide range of fields associated with both library and clone records. Notably, users searching for clones with specific content may enter features (i.e. gene names) or sequence coordinates as search terms. The complete set of indexed fields is documented on the Clone DB website (http://www.ncbi.nlm.nih.gov/clone/content/help/). Queries using combinations of these fields may be entered directly into the search box or users may use the ‘Advanced’ option to create complex queries by selecting specific queries from lists for each of the indexed fields. Alternatively, users may engage the ‘Limits’ option to restrict their searches to a collection of pre-selected, commonly used fields (Figure 2). Search results are returned in a tabular format, where each row provides a feature summary for a specific library or clone record result. These summaries include links to the records themselves.
Figure 2.
Limits facilitate searches of Clone DB. Top: screen shot of the header found on all Clone DB web pages. Complex text-based queries can be directly entered into the search box. Beneath the box, the ‘Limits’ and ‘Advanced’ options provide links to other pages that provide alternate approaches to querying the database. Bottom (red arrow): screen shot of ‘Limits’ page for Clone DB. Users may restrict their searches by selecting from a pre-defined set of commonly used Clone DB search terms for several indexed fields. To further refine searches, users may also combine selected limits with queries entered into the text box.
Library browsers
As a complement to Entrez searches, Clone DB has browsers for its collections of cell-based and genomic libraries. The browsers, which are sortable tables, can be accessed via links on the Clone DB homepage and are a useful mechanism to obtain an overview of the libraries represented in Clone DB. Filters for these browsers enable users to restrict the libraries displayed and navigate to specific library records. Each row of the browsers provides summary data for a specific library record and a link to the corresponding library record display page. For cell-based libraries, filters are provided for library and allele type, library group and creator, as well as the number of clones in the library that have records in Clone DB. For genomic clone libraries, the browser display can be filtered by organism, vector type, distributor and numbers of end or insert sequences associated with the library in Clone DB. Browser-specific FAQ pages for the genomic (http://www.ncbi.nlm.nih.gov/clone/content/browserfaq/) and cell-based (http://www.ncbi.nlm.nih.gov/clone/content/cbbrowserfaq/) library browsers are provided to assist users with their navigation.
FTP
Several reports from Clone DB are available via its public FTP site (ftp://ftp.ncbi.nih.gov/repository/clone/). Within the ‘reports’ directory, data is organized by species. Table 1 lists the reports available and provides a brief description of their content. The Clone DB FTP site also provides a Perl script that enables users to download sequences for genomic clones in bulk. Located in the ‘utility’ directory, the script endseq_dp.pl enables a user to provide a series of end sequence identifiers (available from the Clone DB FTP reports) as input. It returns a file containing the corresponding FASTA sequences, which can then be used for further analyses.
Table 1.
Clone DB FTP reports
| FTP report name | Description |
|---|---|
| clone_acstate_*.out | Per-organism report providing information about insert sequences associated with genomic clone records in Clone DB. Updated weekly. |
| endinfo_*.out | Per-organism report providing information for end sequences associated with genomic clone records in Clone DB. Updated weekly. |
| library_*.out | Per-organism report providing summary information for library records in Clone DB. Updated weekly. |
| end_placement_report_*.out | Per-organism report providing summary information about end sequence placements generated by Clone DB. Updated intermittently. |
| clone_placement_report_*.out | Per-organism report providing summary information about genomic clone placements generated by Clone DB. Updated intermittently. |
*Refers to the NCBI taxonomic identifier for each organism.
Viewing data
Library records
Users can find information about specific genomic and cell-based libraries on individual library display pages. A ‘Summary’ section at the top of each library record page provides an at-a-glance view of key library attributes. For genomic libraries, these attributes include the library name and abbreviation used by Clone DB, the organism from which the library is derived, the names and contact information of the library distributors, the vector type and several calculated attributes, including the total number of corresponding clone records, counts of end and insert sequences and the number of clones that have sequence records for both clone ends. Below the summary, additional library information is provided in a series of tabbed displays. This section provides a detailed description of the library to assist users in determining whether clones from the library are suitable for their research needs, as well information to facilitate their use in a research setting. Each tabbed section represents a set of related library attributes provided in a tabular layout, within which each table row provides data specific to a library segment. For genomic clone libraries, there are tabbed sections that give details about the DNA source, library construction, library statistics, alternate library names and a curated description of the library. A representative genomic record display can be viewed at http://www.ncbi.nlm.nih.gov/clone/library/genomic/260/. Users should note that a library’s display page only includes the tabbed sections and corresponding details available for that library record. For a complete list of genomic library attributes that may be displayed, see http://www.ncbi.nlm.nih.gov/clone/content/overview/.
For cell-based libraries, the library summary fields also provide the library name and organism of origin. Cell-based library record summaries additionally include the allele and library types represented, as well as the library group name. Calculated attributes for cell-based libraries include the total number of library clones represented in Clone DB and the count of associated sequences. Beneath the summary section, tabbed displays provide library construction and library host details. For a representative example of a cell-based library record, see http://www.ncbi.nlm.nih.gov/clone/library/cellbased/828/.
All library records also include a series of ‘Discovery’ links on the right-hand side of each page. These give access to other NCBI and external sites where users may find tools or resources related to clone libraries. Of particular note is the link titled ‘Clones in this library’, which returns the results of an Entrez query of Clone DB for the complete list of associated clone records. Users can find more information about library records at the Clone DB Help page (http://www.ncbi.nlm.nih.gov/clone/content/help/).
Clone records
Clone DB also provides display pages for individual clone records. Like the library record pages, a summary at the top of all individual clone record pages gives an overview of important clone attributes, including the organism, the library to which the clone belongs (with a link to the corresponding library record) and the library type. A series of ‘Discovery’ links at the right hand of each clone record page, like those found on the library record pages, directs users to additional NCBI and external websites that may be of particular interest with respect to clone resources. Displays for cell-based clone records include two tabbed sections beneath the clone summary that provide details about the specific allele represented by the clone and the clone’s distribution (A representative cell-based record is found at http://www.ncbi.nlm.nih.gov/clone/25120061/). The ‘Allele Info’ tab provides the name and type of the allele specific to the clone, as well as links to the associated allele and gene records at MGI, the source of all murine cell-based clone data in Clone DB. This section also provides a link to the corresponding gene record in the NCBI Gene database (1), where users may find additional information about the sequence represented in the clone. For murine cell-based clone records, the ‘Distributors’ tab provides users with information about how to obtain clones from the International Mouse Strain Resource [IMSR, (24)]. This section reports the identities of IMSR centers that supply the allele specifically represented by the clone, as well as other related alleles, either as ES cells or in other biological formats. Links to the IMSR ordering website are also provided to assist users in obtaining these clones.
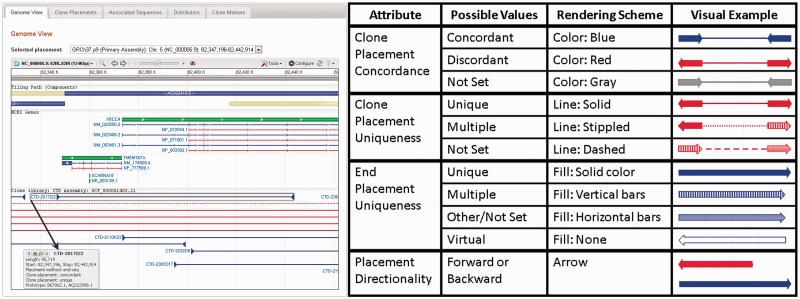
Individual genomic clone records also provide a number of clone-specific details. If NCBI has calculated a placement for the clone on a genome assembly, an ideogram representation of that genome, along with the chromosomal locations at which the clone has been placed, will be displayed to the right of the clone summary (For a representative example, see, http://www.ncbi.nlm.nih.gov/clone/5429/). Below this, the tabbed ‘Genome View’ section utilizes the NCBI Sequence Viewer to display graphical representations of the clone’s NCBI-calculated placements in the context of assembly components, NCBI gene annotation and the placements of other clones belonging to the same library (Figure 3). A legend describing the visualization scheme used to represent clone placement attributes such as concordancy, uniqueness and directionality in the Sequence Viewer is available on the Clone DB clone placements FAQ page http://www.ncbi.nlm.nih.gov/clone/content/placements/. Notably, user-supplied data can be uploaded to the Sequence Viewer instance for visualization in context with the clone placements. In instances where a clone has been placed on multiple assemblies, or at multiple locations within the same assembly, a drop-down menu allows users to selectively display the placement of interest. Holding the mouse over any clone placement in the Sequence Viewer window activates a pop-up window providing details of the clone placement. The ‘Clone Placement’ tab includes a tabular display of all placements associated with the clone. Depending on available data, these include sequence-based placements, such as those calculated by NCBI, as well as non-sequence based (i.e. cytological) placements determined by FISH and other experimental techniques. Placement details provided by Clone DB include the start and stop coordinates and length of the placement, the source of the placement data and where applicable, the concordance and confidence statuses of the placement. Clone DB translates cytological coordinates into sequence based coordinates so that users can compare sequence-based and non-sequence-based placements for consistency.
Figure 3.
Genomic clone placement displays in Clone DB. Left panel: screen shot taken from a genomic clone record display (http://www.ncbi.nlm.nih.gov/clone/5429/). The ‘Genome View’ tab contains a graphic representation of clone placements in the context of assembly features via the NCBI Sequence Viewer. Blue/tan rectangles: assembly components. Green bars: NCBI genes, with RefSeq transcripts and proteins immediately below. At this zoom level, clone placements appear as a series of horizontal lines with triangular ends. The selected placement for the clone of record appears first and is highlighted with a gray box. Additional placements represent other clones from the same genomic library. The black arrow points to the pop-up menu for the selected placement. Right panel: legend for genomic clone placement displays in NCBI Sequence Viewer (legend also found at: http://www.ncbi.nlm.nih.gov/clone/content/placements/).
Other tabs on the genomic clone record pages provide further information about the clone. The data presented in the ‘Associated Sequences’ tab identifies the insert and end sequences associated with that clone, along with links to the corresponding sequence records and details such as sequence length, finishing status and projects in the NCBI BioProjects database (25) with which they may be associated. The ‘Distributors’ tab provides the names of any distributors for the clone and/or its associated library. For clones having fingerprint data in Clone DB, the ‘Fingerprint Info’ tab provides the name of the fingerprint contig in which the clone resides and the number of fingerprint bands and their summed length, as well as the type and status of the clone within the fingerprint contig. Users can find clones for which fingerprint data is available by performing a search that includes the indexed term ‘Has Fingerprint’. For clones on which genomic markers, such as STS, have been placed, the ‘Clone Markers’ tab lists the names and types of such markers, the method by which they were mapped to the clone and the individual/organization that performed the mapping. Together, these sections serve as an integrated resource for clone-associated data that provides users with the information necessary to identify and obtain clones best suited to their research needs. Detailed descriptions of the clone record pages can be found on the Clone DB Help page (http://www.ncbi.nlm.nih.gov/clone/content/help/).
Clone finder
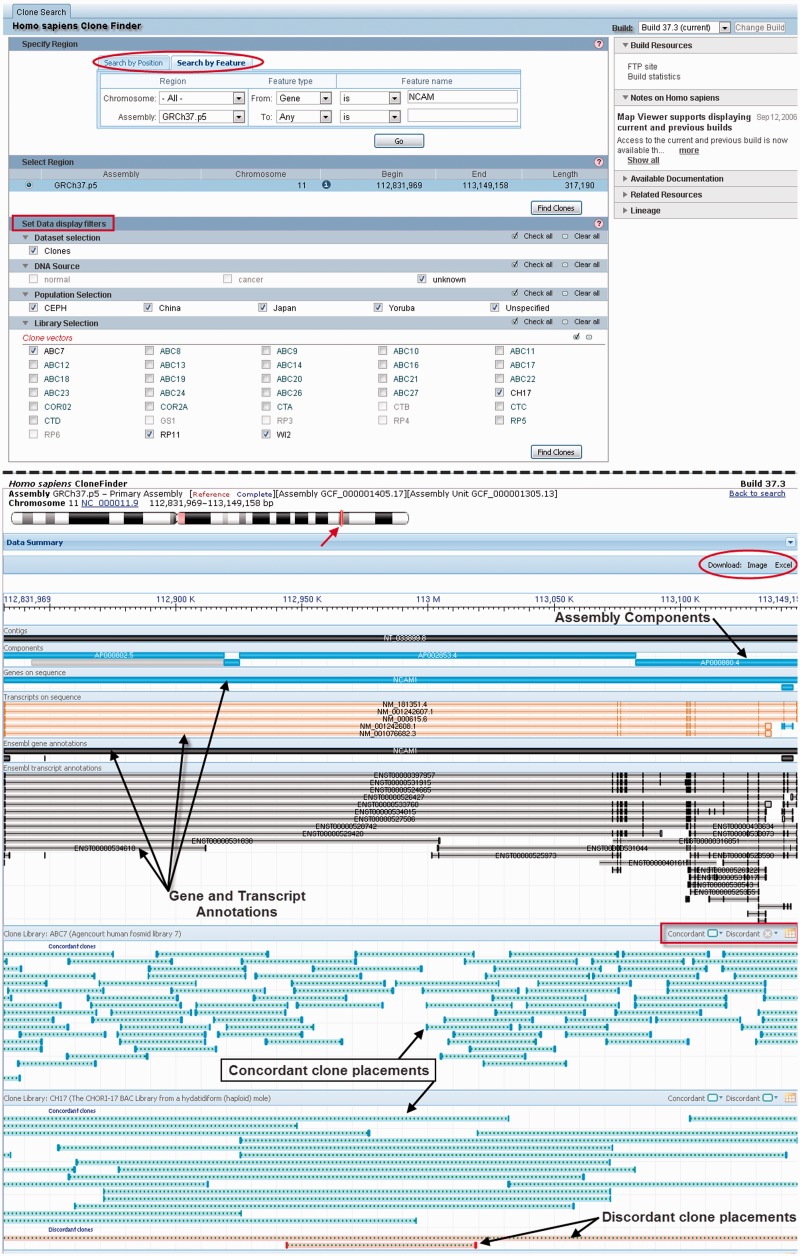
The individual clone record displays described above permit users to view placements and other details for specified genomic clones. A complementary resource, the NCBI Clone Finder, enables users to identify clones placed in specific genomic regions. This tool is particularly useful for finding placed genomic clones that contain a known sequence, gene or other biological feature. Clone Finder can be accessed from a link on the Clone DB homepage. Upon selecting an organism of interest, users may search for clones either by genomic location or by features such as genes, transcripts, SNPs, clone names or markers (i.e. STS). Filters enable users to restrict their search results by attributes such as the clone library, vector type or DNA source. In contrast to the placement displays provided in individual clone records, which only display placements from a single library, Clone Finder can simultaneously show the placements of clones belonging to different libraries. Clone Finder results are displayed graphically in the context of the genome assembly and NCBI and Ensembl (26) gene annotations, with concordant and discordant placements distinguished from one another (Figure 4).The results, including placement details, are also provided in tabular format and can be downloaded as Excel files for further analysis. Clone Finder, together with Clone DB, serve as two resources that enable users to find clones best suited to their research needs and additionally provide them with the information that they will need to work with these experimental reagents. Documentation with detailed instructions for use of Clone Finder is available on the web (http://www.ncbi.nlm.nih.gov/projects/mapview/static/clonefinder_documentation.shtml).
Figure 4.
Clone Finder search results. Top: search interface. Users can query a selected assembly by coordinate or feature (circle). A series of filters (red box) can be applied to restrict search results. This example shows a search for clones belonging to selected libraries that are mapped to the genomic interval containing NCAM1 in the GRCh37.p5 human reference assembly. Bottom: search results. The ideogram at top provides a chromosomal context for the search (red arrow). The graphic display below shows the chromosome coordinates and sequence identifiers of the contigs and components that comprise it, along with annotated RefSeq and Ensembl genes and transcripts (black arrows). Beneath this, placements from each library are presented in separated panels. As noted, concordant placements for clones within the region are colored in green and discordant placements in red. The display of these placements, like the option to the display results in tabular format, can be toggled on and off (red rectangle). Search results can also be downloaded to Excel for further analysis (red circle).
CONCLUSIONS AND FUTURE DEVELOPMENTS
Vector and cell-based clones continue to play important roles in biological research, but users have historically found it difficult to synthesize the wealth of available clone-associated data because it has only been available in resource-specific databases. Clone DB addresses this need and serves as centralized resource that provides integrated information about clones and how researchers may obtain them. Today’s Clone DB represents an expansion and update to the former NCBI Clone Registry. Not only have the number of organisms and libraries represented in the database grown, but so has the amount of information associated with these research reagents.
Work is on-going to further enhance Clone DB. Notably, several efforts are currently underway to improve genomic clone placements. The placement algorithms are being updated to incorporate the use of insert sequences when available, and the Clone Finder tool is undergoing technical changes that will provide users with increased control in searches and display of placement results. While Clone DB currently only contains records for genomic and cell-based clones, the eventual addition of cDNA clone records to the database was planned during its redesign, and it is anticipated that such records will be included in the future. The storage and display of distributor information available in Clone DB is also being improved. Lastly, continuing efforts to further integrate Clone DB with other NCBI databases should improve user access to clone-related data at NCBI in general. Clone DB welcomes user requests for the representation of additional organisms and library and clone records, as well as general user feedback (clonereg-admin@ncbi.nlm.nih.gov).
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Greg Schuler for his helpful insight and discussions of the Clone Registry and suggestions associated with clone name regularization and clone placements. In addition, the authors would like to acknowledge Lukas Wagner and Wonhee Jang for their assistance with issues related to clone name regularization and clone placements.
REFERENCES
- 1.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, Adelson DL, Eichler EE, Elnitski L, Guigo R, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–616. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 6.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura M, Catacchio CR, Alkan C, Marques-Bonet T, Sajjadian S, Graves TA, Hormozdiari F, Navarro A, Malig M, Baker C, et al. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res. 2011;21:1640–1649. doi: 10.1101/gr.124461.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safar J, Bartos J, Janda J, Bellec A, Kubalakova M, Valarik M, Pateyron S, Weiserova J, Tuskova R, Cihalikova J, et al. Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J. Cell Mol. Biol. 2004;39:960–968. doi: 10.1111/j.1365-313X.2004.02179.x. [DOI] [PubMed] [Google Scholar]
- 9.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babiychuk E, Fuangthong M, Van Montagu M, Inze D, Kushnir S. Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl Acad. Sci. USA. 1997;94:12722–12727. doi: 10.1073/pnas.94.23.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 2007;63:351–364. doi: 10.1007/s11103-006-9093-z. [DOI] [PubMed] [Google Scholar]
- 12.Lukacsovich T, Yamamoto D. Trap a gene and find out its function: toward functional genomics in Drosophila. J. Neurogenet. 2001;15:147–168. doi: 10.3109/01677060109167373. [DOI] [PubMed] [Google Scholar]
- 13.Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Burger A, Bult CJ, et al. The mammalian gene function resource: the international knockout mouse consortium. Mamm. Genome Official J. Int. Mamm. Genome Soc. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skarnes WC, von Melchner H, Wurst W, Hicks G, Nord AS, Cox T, Young SG, Ruiz P, Soriano P, Tessier-Lavigne M, et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen GM, Markesich DC, Burnett MB, Zhu Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP, Abuin A. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008;18:1670–1679. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fjell CD, Bosdet I, Schein JE, Jones SJ, Marra MA. Internet Contig Explorer (iCE)–a tool for visualizing clone fingerprint maps. Genome Res. 2003;13:1244–1249. doi: 10.1101/gr.819303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, Kim UJ, Kuo WL, Olivier M, et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang W, Yonescu R, Knutsen T, Brown T, Reppert T, Sirotkin K, Schuler GD, Ried T, Kirsch IR. Linking the human cytogenetic map with nucleotide sequence: the CCAP clone set. Cancer Genet. Cytogenet. 2006;168:89–97. doi: 10.1016/j.cancergencyto.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Kadin JA, Bult CJ, Eppig JT, Oakley DJ, Briois S, et al. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39:D849–D855. doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgulis A, Gertz EM, Schaffer AA, Agarwala R. WindowMasker: window-based masker for sequenced genomes. Bioinformatics. 2006;22:134–141. doi: 10.1093/bioinformatics/bti774. [DOI] [PubMed] [Google Scholar]
- 24.Eppig JT, Strivens M. Finding a mouse: the International Mouse Strain Resource (IMSR) Trends Genet. 1999;15:81–82. doi: 10.1016/s0168-9525(98)01665-5. [DOI] [PubMed] [Google Scholar]
- 25.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]