Abstract
HAMAP (High-quality Automated and Manual Annotation of Proteins—available at http://hamap.expasy.org/) is a system for the classification and annotation of protein sequences. It consists of a collection of manually curated family profiles for protein classification, and associated annotation rules that specify annotations that apply to family members. HAMAP was originally developed to support the manual curation of UniProtKB/Swiss-Prot records describing microbial proteins. Here we describe new developments in HAMAP, including the extension of HAMAP to eukaryotic proteins, the use of HAMAP in the automated annotation of UniProtKB/TrEMBL, providing high-quality annotation for millions of protein sequences, and the future integration of HAMAP into a unified system for UniProtKB annotation, UniRule. HAMAP is continuously updated by expert curators with new family profiles and annotation rules as new protein families are characterized. The collection of HAMAP family classification profiles and annotation rules can be browsed and viewed on the HAMAP website, which also provides an interface to scan user sequences against HAMAP profiles.
INTRODUCTION
Falling costs and continuing technological improvements mean that genome sequencing has become a routine tool in life science research. The availability of thousands of finished genome sequences covering taxonomic ranges from individual strains to whole kingdoms has allowed biologists to ask new questions about the evolution of individual proteins, genomes and even species (1). Annotated genomes also provide an essential starting point in the construction of genome-scale models of cellular processes, particularly of cellular metabolism (2). These models may in turn serve as a framework for the iterative enhancement of genome annotation, providing contextual information that is complementary to the primary sequence and that can be used to infer potential new functions for uncharacterized genes (3). These and other applications are critically dependent on the quality of genome annotation, both of the predicted gene models, and of the functional assignments that are made to the putative gene products.
Genome sequencing technologies are now within the reach of many individual research groups, meaning that the pace of data production, and subsequent submission to archival resources such as the International Nucleotide Sequence Database Collaboration (INSDC; composed of GenBank, the European Nucleotide Archive and the DNA Data Bank of Japan) (4) is unlikely to slow. Exploiting this data requires genome annotation that is as complete and accurate as possible, but providing this annotation remains a challenge. The development of shared standard operating procedures by the major sequencing centers (5) will undoubtedly improve the quality of the resulting archival annotations. These may be further enhanced by the provision of detailed functional annotation by third-party resources that can be updated on a regular basis as new knowledge becomes available.
One source of such annotation is the UniProt Knowledgebase, UniProtKB, a resource of protein sequences and associated functional information (6). UniProtKB is composed of two sections: UniProtKB/Swiss-Prot, which includes records that have been manually reviewed and curated by a human curator, and UniProtKB/TrEMBL, which includes unreviewed records. UniProtKB sequences from both sections are classified by InterPro (7), which groups signatures for the identification of conserved protein domains and families from a number of resources, and which also provides functional annotation in the form of curated terms from the Gene Ontology (GO) (8,9). The InterPro classification has been exploited for the construction of annotation rules that link InterPro signatures and other information to relevant functional annotation from UniProtKB/Swiss-Prot (10–12). Other resources providing functional annotation include KEGG (13), MetaCyc (14) and the SEED (15), which combine curated reference data on metabolism with methods to ‘project’ this data to new genomes. In the case of KEGG and the SEED, functions are inferred based on sequence homology, whereas the MetaCyc PathoLogic algorithm makes ‘chained’ inferences based on annotations in INSDC records (16). Another useful source of annotation for enzymes is PRIAM, which automatically identifies conserved sequence signatures in annotated enzymes from UniProtKB, and uses these signatures to identify and annotate uncharacterized homologs (17).
Genome sequencing centers and other users rely on the information available in these and other systems to annotate new genomes and proteins. To enhance the provision of such information in UniProtKB, we previously developed the HAMAP system (for High-quality Automated and Manual Annotation of Proteins) (18). HAMAP was originally designed to annotate protein sequences from prokaryotic species to the quality standards required by UniProtKB/Swiss-Prot, exactly as a human curator would do, and was used in the construction and development of UniProtKB/Swiss-Prot (18). HAMAP is based on a collection of manually curated family profiles, which are used to determine family membership of protein sequences. HAMAP profiles are linked to manually curated annotation rules, which specify the annotation that can be applied to members of the protein family, and which include additional control statements that supervise the propagation of this annotation to member sequences. In the remainder of this article, we describe the current status and new developments in HAMAP, and briefly describe how HAMAP will be used to annotate UniProtKB in the future.
HAMAP: A COLLECTION OF MANUALLY CURATED FAMILY PROFILES WITH ASSOCIATED ANNOTATION RULES
HAMAP family profiles
HAMAP family profiles are used to determine family membership of protein sequences. HAMAP profiles are automatically generated from manually curated seed alignments of trusted family member sequences. This set of trusted member sequences normally includes all characterized family members from UniProtKB/Swiss-Prot, plus a representative selection of other sequences that provide broad taxonomic coverage of the target family. Sequences are selected using iterative and reciprocal BLAST searches (19), and the resulting sets are compared with those from other resources of protein families and homologs including HOGENOM (20), OrthoDB (21), TIGRFAMs (22), Pfam (23) and PROSITE (24). All protein sequences that are included in the seed alignment are manually checked, and where necessary corrected. This may typically involve rectification of erroneous start sites or erroneous gene model predictions. These corrections are subsequently integrated into UniProtKB/Swiss-Prot, thereby guaranteeing that the corrected sequences remain fixed and synchronized with the HAMAP family profiles of which they are a member.
Following the automatic generation of a detection profile from the seed alignment (25), the profile is calibrated using the standard PROSITE procedure (26). The profile is scanned against a database of randomized protein sequences from UniProtKB, and the parameters of an extreme value distribution are estimated from the score distribution obtained (26). These parameters are subsequently used in the normalization of the raw scores using an affine transformation (26). The normalized scores are related to the commonly used E-value, which is the expected number of matches with a score equal to or greater than a given score that would be expected to arise by chance. For example, a match with a normalized score of 9.0 would be expected to occur roughly once in a database of one billion residues.
During profile construction and calibration, all matches to the profile are extracted from UniProtKB and the lowest scoring member sequence of the seed alignment is used to define an initial threshold value (or trusted cutoff score) for the normalized scores to each profile. Curators can manually adjust this cutoff to include lower scoring member sequences, or raise it to reduce the possibility of false positive matches. Curators may also choose to alter the composition of the original seed alignment to enhance the specificity of the profile, performing iterative profile searches until a satisfactory score distribution is obtained.
HAMAP annotation rules
Each HAMAP family profile may be associated with one or more HAMAP annotation rules. When multiple rules are associated to a single profile, then each rule will normally apply to a distinct taxonomic group. HAMAP annotation rules define the relevant annotations for protein sequences that match the associated HAMAP profile, and are manually created using information from UniProtKB/Swiss-Prot entries. Annotations are provided in the form of free text, controlled vocabularies from UniProtKB, such as UniPathway (27), and terms from the GO (9). Typical annotations may describe protein function, enzymatic activities, subcellular location, and pathway membership, as well as specific sequence features such as active sites and ligand-binding residues. Annotations may be subject to control statements that limit their propagation to only those sequences satisfying one or more conditions, such as a requirement for the presence of specific conserved functional residues (18).
RECENT DEVELOPMENTS IN HAMAP
Automatic annotation of UniProtKB/TrEMBL
HAMAP was originally developed as a tool for the annotation of microbial protein sequences to the same level of detail and to the same quality standards as manually curated UniProtKB/Swiss-Prot records (18). HAMAP was used to annotate UniProtKB/TrEMBL records, which were then carefully checked and integrated into UniProtKB/Swiss-Prot. Since our last publication in 2009 describing the HAMAP classification and annotation system, we have made significant alterations to the way that HAMAP is used during the UniProtKB curation and production process. HAMAP family profiles have now been integrated into InterPro, and HAMAP rule-based annotation is now applied in a fully automated fashion to UniProtKB/TrEMBL records. Rules and conditions are interpreted in precisely the same way as before, and conditional annotations are applied only to those proteins that satisfy the relevant criteria. The set of HAMAP rules is also being combined with annotation rules from RuleBase (11,12) and PIR (28) into a single automatic annotation system for UniProtKB/TrEMBL, UniRule, which will be the subject of a forthcoming publication by the UniProt consortium. Although HAMAP rules will be part of a larger integrated UniRule system, we will continue to maintain the HAMAP protein family profiles as a basis for protein classification and rule-based annotation within UniRule. Together, these developments will help leverage the experimental annotation and manual curation effort from UniProtKB/Swiss-Prot into UniProtKB/TrEMBL, providing functional annotation for sequences for which no experimental data exists.
Extension of HAMAP to eukaryotes
The original scope of the HAMAP system was largely determined by the taxonomic distribution of the complete genomes that were available at the time of its inception. As more genomes from other taxonomic groups such as eukaryotes have become available in UniProtKB (6), through pipelines importing sequences from resources such as Ensembl (29), we have begun to observe an ever-increasing number of matches to existing HAMAP families in these genomes. We have therefore extended the scope of HAMAP families and annotation rules to include proteins from eukaryotic species, and annotations derived from these rules have been available in UniProtKB since UniProt release 2012_09 of October 2012.
Updates to the website
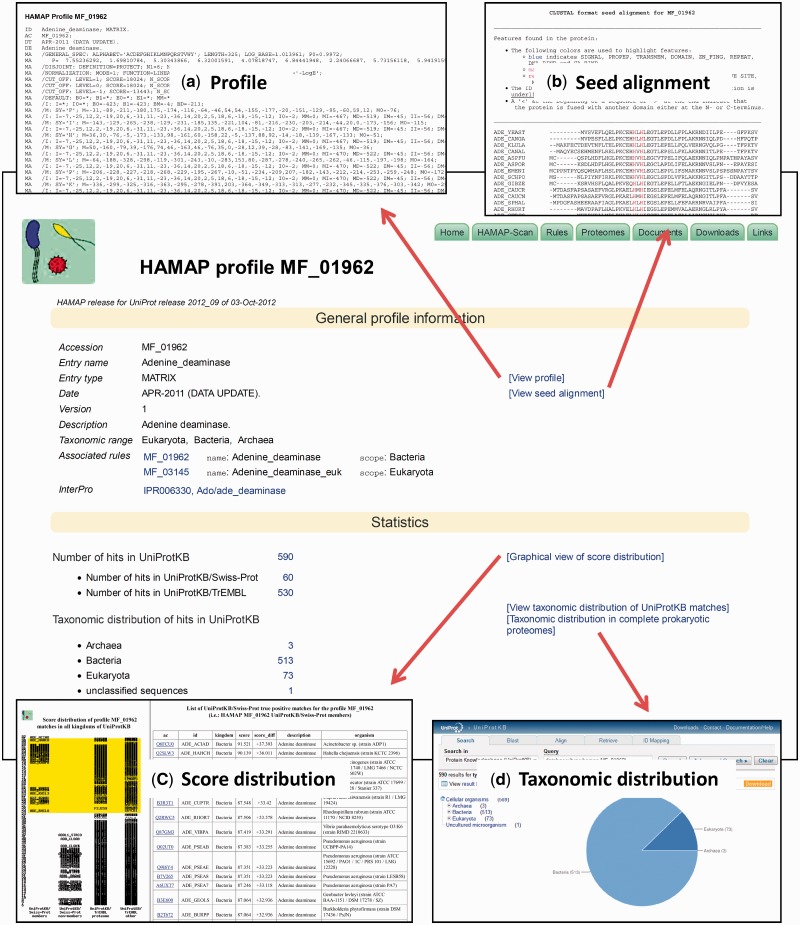
HAMAP family profiles and their associated annotation rules are made available as independent pages on the HAMAP website. As more than one annotation rule can be triggered by a single HAMAP family profile, each rule is assigned a distinct page, and each of these is linked to the ‘trigger’ profile. A typical HAMAP profile page provides, in addition to the profile itself, relevant information such as a family name and description, taxonomic range (as a list of matching superkingdoms), associated annotation rule(s) and cross-references to InterPro, as well as information on the score distribution of matching proteins, including those that fall below the trusted cutoff (Figure 1). In line with these changes, we have also redesigned the web view of the annotation rules and added new options for searching and accessing the collection of annotation rules. As well as listing all rules by taxonomic scope, enzyme class, pathway, feature key or keywords, it is now also possible to browse the annotation rules by GO terms. These GO annotations are also available for download on the UniProt-GO Annotation database ftp site (see ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/UNIPROT/).
Figure 1.
A sample HAMAP profile page. The page provides information such as a family name and description, taxonomic range of the hits, associated annotation rule(s), cross-references to InterPro and access to matching proteins in UniProtKB. Additionally, links on the page provide access to (a) the actual family classification profile, (b) the seed alignment that was used to generate the profile with highlighted features from the annotation rule, (c) an interactive, graphical view of the score distribution of matching proteins, including those that fall below the trusted cutoff, and (d) an expandable view of the taxonomic distribution of matching proteins in UniProtKB.
HAMAP STATISTICS AND AVAILABILITY
As of release 2012_08 of UniProt, HAMAP contains 1780 family classification profiles and 1720 annotation rules. The family profiles cover 2 317 216 UniProtKB entries, which is close to 10% of all sequences in UniProtKB. Considering only the 1696 complete prokaryotic proteomes of UniProtKB, the coverage of HAMAP is around 14% of an ‘average’ prokaryotic proteome. The precise figure may vary considerably depending on our knowledge of the organism, the degree to which it has been studied, and the size of its genome, being around 25% for the model organism Escherichia coli, and reaching 64% for the reduced genome of Buchnera aphidicola. Coverage is dependent on the number of available rules, and we are continuing to add new profiles and rules to further improve the coverage of proteins by the HAMAP system. While HAMAP annotations are made available through UniProtKB, HAMAP family profiles and rules can also be used directly for the annotation of protein sequences through our web interface at http://hamap.expasy.org/hamap_scan.html. Users may submit individual protein sequences or complete microbial proteomes to be scanned against the entire collection of HAMAP profiles and annotated by HAMAP rules.
CONCLUDING REMARKS
We describe the extension of the scope of the HAMAP system of family classification and annotation to eukaryotic proteins and its application in the fully automatic annotation of the unreviewed section of the UniProt knowledgebase, UniProtKB/TrEMBL. These changes were implemented without compromising the quality of the annotations produced, which remains equal to that of manually curated UniProtKB/Swiss-Prot records. HAMAP annotation rules include numerous checks (or conditions) that must be satisfied for annotation propagation to proceed, ensuring high specificity of the annotations produced. This design feature is intended to reduce the likelihood of over-annotation, a relatively common error in some automated pipelines (30). In the near future, the HAMAP annotation rules will be made available as one element of an integrated system of automatic annotation for UniProtKB/TrEMBL, UniRule. This will be described in a future publication by the UniProt consortium. In the context of UniRule, we will continue to maintain the HAMAP protein family profiles as a basis for protein classification and the development of new annotation rules as new functions are discovered.
FUNDING
UniProt is mainly supported by the National Institutes of Health (NIH) [1 U41 HG006104-03]. Additional support for the EBI’s involvement in UniProt comes from the NIH [2P41 HG02273] and the British Heart Foundation [SP/07/007/23671]. Swiss-Prot activities at the SIB are supported by the Swiss Federal Government through the Federal Office of Education and Science and the European Commission contracts SLING [226073], Gen2Phen [200754] and MICROME [222886]. PIR’s UniProt activities are also supported by the NIH [5R01GM080646-07, 3R01GM080646-07S1, 5G08LM010720-03, and 8P20GM103446-12], and the National Science Foundation (NSF) [DBI-1062520]. Page charges for this article were paid by the Swiss Federal Government through the Federal Office of Education and Science. Funding for open access charge: Swiss Federal Government through the Federal Office of Education and Science.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
UniProt has been prepared by: Rolf Apweiler, Maria Jesus Martin, Claire O’Donovan, Michele Magrane, Yasmin Alam-Faruque, Emanuela Alpi, Ricardo Antunes, Joanna Arganiska, Elisabet Barrera Casanova, Benoit Bely, Mark Bingley, Carlos Bonilla, Ramona Britto, Borisas Bursteinas, Wei Mun Chan, Gayatri Chavali, Elena Cibrian-Uhalte, Alan Da Silva, Maurizio De Giorgi, Emily Dimmer, Francesco Fazzini, Paul Gane, Alexander Fedotov, Leyla Garcia Castro, Penelope Garmiri, Emma Hatton-Ellis, Reija Hieta, Rachael Huntley, Julius Jacobsen, Rachel Jones, Duncan Legge, Wudong Liu, Jie Luo, Alistair MacDougall, Prudence Mutowo, Andrew Nightingale, Sandra Orchard, Samuel Patient, Klemens Pichler, Diego Poggioli, Sangya Pundir, Luis Pureza, Guoying Qi, Steven Rosanoff, Tony Sawford, Harminder Sehra, Edward Turner, Vladimir Volynkin, Tony Wardell, Xavier Watkins, Hermann Zellner, Matt Corbett, Mike Donnelly, Pieter van Rensburg, Mickael Goujon, Hamish McWilliam, and Rodrigo Lopez at the European Bioinformatics Institute (EBI). Ioannis Xenarios, Lydie Bougueleret, Alan Bridge, Sylvain Poux, Nicole Redaschi, Andrea Auchincloss, Kristian Axelsen, Parit Bansal, Delphine Baratin, Pierre-Alain Binz, Marie-Claude Blatter, Brigitte Boeckmann, Jerven Bolleman, Emmanuel Boutet, Lionel Breuza, Alan Bridge, Edouard de Castro, Lorenzo Cerutti, Elisabeth Coudert, Beatrice Cuche, Mikael Doche, Dolnide Dornevil, Severine Duvaud, Anne Estreicher, Livia Famiglietti, Marc Feuermann, Elisabeth Gasteiger, Sebastien Gehant, Vivienne Gerritsen, Arnaud Gos, Nadine Gruaz-Gumowski, Ursula Hinz, Chantal Hulo, Janet James, Florence Jungo, Guillaume Keller, Vicente Lara, Philippe Lemercier, Jocelyne Lew, Damien Lieberherr, Xavier Martin, Patrick Masson, Anne Morgat, Teresa Neto, Salvo Paesano, Ivo Pedruzzi, Sandrine Pilbout, Monica Pozzato, Manuela Pruess, Catherine Rivoire, Bernd Roechert, Michel Schneider, Christian Sigrist, Karin Sonesson, Sylvie Staehli, Andre Stutz, Shyamala Sundaram, Michael Tognolli, Laure Verbregue, Anne-Lise Veuthey, and Mohamed Zerara at the Swiss Institute of Bioinformatics (SIB). Cathy H. Wu, Cecilia N. Arighi, Leslie Arminski, Chuming Chen, Yongxing Chen, Hongzhan Huang, Abhishek Kukreja, Kati Laiho, Peter McGarvey, Darren A. Natale, Thanemozhi G. Natarajan, Natalia V. Roberts, Baris E. Suzek, C. R. Vinayaka, Qinghua Wang, Yuqi Wang, Lai-Su Yeh, Meher Shruti Yerramalla, and Jian Zhang at the Protein Information Resource (PIR).
REFERENCES
- 1.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat. Rev. Genet. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 3.Orth JD, Palsson BO. Systematizing the generation of missing metabolic knowledge. Biotechnol. Bioeng. 2010;107:403–412. doi: 10.1002/bit.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karsch-Mizrachi I, Nakamura Y, Cochrane G. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40:D33–D37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity G, Kodira CD, Kyrpides N, Madupu R, Markowitz V, et al. Toward an online repository of Standard Operating Procedures (SOPs) for (meta)genomic annotation. OMICS. 2008;12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O'Donovan C, Martin MJ, Bely B, Browne P, Mun Chan W, Eberhardt R, et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2012;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gene Ontology Consortium. The Gene Ontology: enhancements for 2011. Nucleic Acids Res. 2012;40:D559–D564. doi: 10.1093/nar/gkr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kretschmann E, Fleischmann W, Apweiler R. Automatic rule generation for protein annotation with the C4.5 data mining algorithm applied on SWISS-PROT. Bioinformatics. 2001;17:920–926. doi: 10.1093/bioinformatics/17.10.920. [DOI] [PubMed] [Google Scholar]
- 11.Biswas M, O'Rourke JF, Camon E, Fraser G, Kanapin A, Karavidopoulou Y, Kersey P, Kriventseva E, Mittard V, Mulder N, et al. Applications of InterPro in protein annotation and genome analysis. Brief Bioinform. 2002;3:285–295. doi: 10.1093/bib/3.3.285. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann W, Moller S, Gateau A, Apweiler R. A novel method for automatic functional annotation of proteins. Bioinformatics. 1999;15:228–233. doi: 10.1093/bioinformatics/15.3.228. [DOI] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeJongh M, Formsma K, Boillot P, Gould J, Rycenga M, Best A. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics. 2007;8:139. doi: 10.1186/1471-2105-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karp PD, Latendresse M, Caspi R. The pathway tools pathway prediction algorithm. Stand. Genomic Sci. 2012;5:424–429. doi: 10.4056/sigs.1794338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Penel S, Arigon AM, Dufayard JF, Sertier AS, Daubin V, Duret L, Gouy M, Perriere G. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009;10(Suppl.6):S3. doi: 10.1186/1471-2105-10-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, Kriventseva EV. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res. 2011;39:D283–D288. doi: 10.1093/nar/gkq930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gattiker A, Michoud K, Rivoire C, Auchincloss AH, Coudert E, Lima T, Kersey P, Pagni M, Sigrist CJ, Lachaize C, et al. Automated annotation of microbial proteomes in SWISS-PROT. Comput. Biol. Chem. 2003;27:49–58. doi: 10.1016/s1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 26.Pagni M, Jongeneel CV. Making sense of score statistics for sequence alignments. Brief Bioinform. 2001;2:51–67. doi: 10.1093/bib/2.1.51. [DOI] [PubMed] [Google Scholar]
- 27.Morgat A, Coissac E, Coudert E, Axelsen KB, Keller G, Bairoch A, Bridge A, Bougueleret L, Xenarios I, Viari A. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Res. 2012;40:D761–D769. doi: 10.1093/nar/gkr1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasudevan S, Vinayaka CR, Natale DA, Huang H, Kahsay RY, Wu CH. Structure-guided rule-based annotation of protein functional sites in UniProt knowledgebase. Methods Mol. Biol. 2011;694:91–105. doi: 10.1007/978-1-60761-977-2_7. [DOI] [PubMed] [Google Scholar]
- 29.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]