Abstract
TropGeneDB (http://tropgenedb.cirad.fr) was created to store genetic, molecular and phenotypic data on tropical crop species. The most common data stored in TropGeneDB are molecular markers, quantitative trait loci, genetic and physical maps, genetic diversity, phenotypic diversity studies and information on genetic resources (geographic origin, parentage, collection). TropGeneDB is organized on a crop basis with currently nine public modules (banana, cocoa, coconut, coffee, cotton, oil palm, rice, rubber tree, sugarcane). Crop-specific Web consultation interfaces have been designed to allow quick consultations and personalized complex queries. TropGeneDB is a component of the South Green Bioinformatics Platform (http://southgreen.cirad.fr/).
INTRODUCTION
TropGeneDB is an information system initially developed by CIRAD (French Agricultural Research Centre for International Development) to manage various kinds of data on tropical crops (1).
TropGeneDB can record crop information on:
molecular markers, quantitative trait loci (QTLs), genetic and physical maps
genetic diversity studies
phenotypic diversity studies based on agro-morphological traits or traits measuring susceptibility/resistance to aggression (diseases, salinity, drought)
geographic origin, parentage, collection
The TropGeneDB project was initiated using the ACEDB database management system (2), but we migrated to MySQL to follow the evolution of computer technology. TropGeneDB is organized on a crop basis. Since the first version of TropGeneDB (1) containing three running modules (cocoa, banana and sugarcane), six additional modules have been implemented: coconut, coffee, cotton, oil palm, rice and rubber tree. A sorghum module exists, but it is still in private access.
New crop-specific Web consultation interfaces have been designed to allow quick and complex queries with user-friendly result representation. Links to the GMOD CMAP viewer (the Comparative Map Viewer) (3) have been integrated.
All the data in TropGeneDB are public and generally linked to published scientific articles. Most of the data in TropGeneDB are original data not available in other Web information systems. Indeed, the TropGeneDB rice module stores unique genetic and phenotypic data on European collections of rice (4). The TropGeneDB banana module was identified by the Global Musa Genomics Consortium as a reference database for banana markers and genetic maps (5). No other equivalent database exists for most of our crop modules (coconut, cocoa, rubber tree, oil palm, sugarcane). Our resources contain data about important crops for the agrarian economy of many tropical countries. Indeed, these data can be exploited for rational use of genetic diversity available from germplasm collections, genome mapping and marker-assisted selection.
TropGeneDB is a component of the South Green Bioinformatics Platform (http://southgreen.cirad.fr/) and is accessible at http://tropgenedb.cirad.fr.
DATABASE CONTENTS
Currently, TropGeneDB contains ∼19 800 molecular markers and 9500 germplasm entries (Table 1). Molecular data comprise genotypes at various types of markers (SNP, DArT, SSR, RFLP, AFLP, etc.), information on the markers themselves (probes, primers, sequences, etc.), QTLs, genetic and physical maps. Germplasm entries are linked to passport data (collections, accession identifier, country of origin, etc.) and much other detailed information: taxonomy, ploidy, ecosystem, etc. They are also linked to detailed phenotypic information based on agronomic and morphological traits and measurements of susceptibility or resistance to abiotic and biotic stress. Most of the rice trait data, used in phenotyping and QTL studies, are annotated with trait ontology terms (6).
Table 1.
Number of TropGeneDB entries (15 July 2012)
| Markers | Germplasms | QTLs | Map studies | Phenotyping studies | |
|---|---|---|---|---|---|
| Banana | 1068 | 541 | 5 | ||
| Cocoa | 2108 | 187 | 8 | ||
| Coffee | 258 | 2 | |||
| Coconut | 471 | 174 | 63 | 1 | |
| Cotton | 5786 | 13 | 478 | 8 | |
| Oil palm | 969 | ||||
| Rice | 3466 | 2041 | 2179 | 1 | 26 |
| Rubber tree | 724 | 4 | 3 | ||
| Sugarcane | 5026 | 6737 | 81 | 6 | 33 |
| Total | 19 876 | 9512 | 2988 | 32 | 59 |
WEB CONSULTATION INTERFACE
Query forms
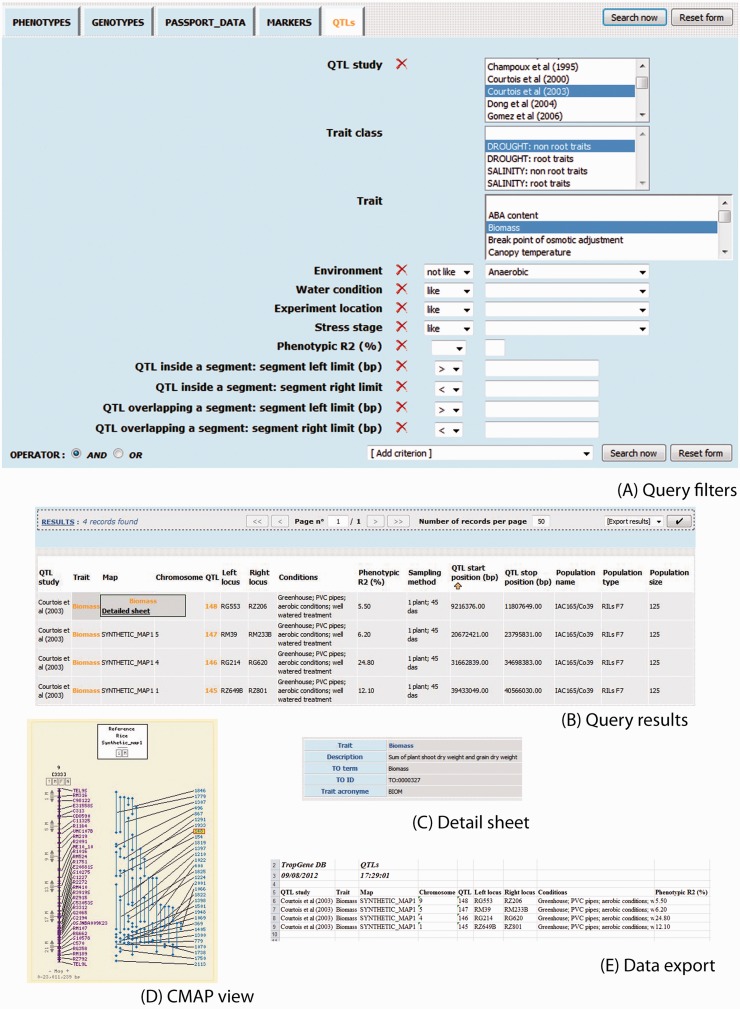
Each crop module has several interfaces to carry out specific requests. The interfaces are tab-based, with each tab containing a different query form that leads to different types of data: phenotypes, genotypes, passport data, markers, QTLs, etc. A query form consists of various kinds of filters represented by standard input widgets. An important point is the ability to select the operator (AND/OR) applied for grouping together the filters defined by the used widgets. Filters corresponding to numerical values can be combined using relational operators (more than, equal to, etc.). For mapping data, users can filter on mapped markers and QTLs inside or overlapping a segment of given left and right limits (bp or cM) (Figure 1A).
Figure 1.
Example of a rice QTL query and corresponding results. (A) Query filters, (B) Query results, (C) Detail sheet, (D) CMAP view, (E) Data export.
Some text boxes have lookup lists tied to them. This is particularly useful when a non-trivial, exact string is expected. Filters where a red cross lies between the criterion label and the widget can be removed by clicking on the indicated cross icon. Those filters can also be (re-)added, several times if needed, using the ‘Add criterion’ list box. Having several identical filters only makes sense when using the OR operator because otherwise selecting two different values for the same criterion would result in an empty data set.
Filters for which no red cross appears primarily consist of list boxes whose contents are subject to a dependency relationship. This kind of relationship ties list boxes two by two: the contents of the second one are automatically updated when selection changes in the first. This hierarchical link can be between a genotyping study and the list of corresponding germplasms, or between a marker type and the corresponding list of markers. Furthermore, for rice phenotype and QTL studies, we have defined a standardized hierarchy of trait classes (Figure 1A).
Result data
Search results are displayed in tables where each column represents a relevant attribute (database field) (Figure 1B). Visualized data can be sorted by any of those fields by clicking on the column header label. In some cases where additional details are available regarding a given attribute, its value is highlighted in orange: when moused-over, an extra layer appears where one or more links can be clicked. Indeed, the link ‘Detail sheet’ provides internal TropGeneDB details on the attribute (Figure 1C). External links are also available like Gramene (6) for the germplasms and markers of the rice TropGeneDB module. For maps, mapped markers and QTLs, links to a GMOD CMAP viewer (the Comparative Map Viewer) (3) have been integrated. The CMAP viewer provides a graphical representation of the correspondences between markers on different genetic and/or physical maps (Figure 1D). From the CMAP viewer, a link is enabled to the GMOD GBROWSE (the genome viewer) when right clicking a mapped object. This link is reciprocal. Pagination is used for displaying the results. Apart from being able to move to the first, next, previous or last page, users can also define the number of records per page, or directly jump to a page by specifying its index.
Exporting results
At the end of the navigation bar can be found an ‘Export results’ list box that lets you select an export format: Excel or Text (csv) (Figure 1E). Depending on the kind of data that are being dealt with, two extra formats may be offered: Excel matrix and Text matrix. Indeed, the results of queries on phenotype or genotype data, which respectively involve traits and germplasms, and markers and germplasms, are in row mode by default, i.e. one row for a combination of germplasms/traits or germplasms/markers. When exported in matrix format, those results are rebuilt as a matrix with the germplasms in rows and the traits or markers in columns.
DATA SUBMISSION
TropGeneDB data have been submitted by CIRAD teams and scientists from other institutions working on tropical crops: a data origin Web page is available for each crop module. People wishing to submit data may download submission templates (Microsoft Excel files), instructions for filling in the templates and can contact us using the Web form. Submitted data quality and integrity are checked by biologists. These biologists are researchers, experts for a given crop, who help the database administrator in checking crucial points: for instance, in standardizing the names of the germplasms, the markers and the traits. They also assess the relevance of the submitted studies.
DESIGN AND IMPLEMENTATION
TropGeneDB is a Web application based on MySQL databases, with one per crop that can be queried using Java customizable interfaces automatically generated to fit the database contents. A generic database model has been developed and is used for all the crop modules.
CONCLUSION
The TropGeneDB database is a long-term project. This database is permanently updated: the range of crops will soon be broaden to alfalfa, einkorn, olive tree, pearl millet, sorghum, tomato. New data types, such as association results and linkage disequilibrium, will also be added.
The user interface could be improved: the input widgets could be placed differently to prevent the need of scrolling up and down to see them all when they are numerous.
AVAILABILITY
Authors who use and download data from TropGeneDB are encouraged to cite this article.
FUNDING
CIRAD (Centre de Coopération Internationale en Recherche Agronomique pour le Développement); International Consortium for Sugarcane Biotechnology (to V.J.); Agropolis Foundation through the Agropolis Resource Centre for Crop Conservation, Adaptation and Diversity project (ARCAD) (to G.S.). Funding for open access charge: CIRAD.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors should like to thank all the expert biologists involved in TropGeneDB data curation. They also thank Gaëtan Droc for the reciprocal links between CMAP and GBROWSE. They are grateful to Peter Biggins for help in English editing. C.H. and V.J. implemented the new version of the MySQL database, developed the data submission templates and the Perl scripts for data insertion. G.S. developed the Java Web interfaces. C.H. managed data updates and the design of the Web query forms. M.R. coordinated the project and wrote the article.
REFERENCES
- 1.Ruiz M, Rouard M, Raboin LM, Lartaud M, Lagoda P, Courtois B. TropGENE-DB, a multi-tropical crop information system. Nucleic Acids Res. 2004;32:D364–D367. doi: 10.1093/nar/gkh105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelley S. Getting started with Acedb. Brief. Bioinform. 2000;1:131–137. doi: 10.1093/bib/1.2.131. [DOI] [PubMed] [Google Scholar]
- 3.Youens-Clark K, Faga B, Yap IV, Stein L, Ware D. CMap 1.01: a comparative mapping application for the Internet. Bioinformatics. 2009;25:3040–3042. doi: 10.1093/bioinformatics/btp458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courtois B, Frouin J, Greco R, Bruschi G, Droc G, Hamelin C, Ruiz M, Clément G, Evrard JC, van Coppenole S, et al. Genetic diversity and population structure in a european collection of rice. Crop Sci. 2012;52:1663–1675. [Google Scholar]
- 5. Rouard,M., Carpentier,S.C., Bocs,S., Droc,G., Argout,X., Roux,N. and Ruiz,M. (2012) In: Pillay Michael,U.G.K.C. (ed.). Science Publishers, Enfield, pp. 194–216.
- 6.Youens-Clark K, Buckler E, Casstevens T, Chen C, Declerck G, Derwent P, Dharmawardhana P, Jaiswal P, Kersey P, Karthikeyan AS, et al. Gramene database in 2010: updates and extensions. Nucleic Acids Res. 2011;39:D1085–D1094. doi: 10.1093/nar/gkq1148. [DOI] [PMC free article] [PubMed] [Google Scholar]