Abstract
Genomicus (http://www.dyogen.ens.fr/genomicus/) is a database and an online tool that allows easy comparative genomic visualization in >150 eukaryote genomes. It provides a way to explore spatial information related to gene organization within and between genomes and temporal relationships related to gene and genome evolution. For the specific vertebrate phylum, it also provides access to ancestral gene order reconstructions and conserved non-coding elements information. We extended the Genomicus database originally dedicated to vertebrate to four new clades, including plants, non-vertebrate metazoa, protists and fungi. This visualization tool allows evolutionary phylogenomics analysis and exploration. Here, we describe the graphical modules of Genomicus and show how it is capable of revealing differential gene loss and gain, segmental or genome duplications and study the evolution of a locus through homology relationships.
INTRODUCTION
Visualization interfaces are critical tools to explore and interpret complex multi-dimensional genomic data. Comparative genomic data are particularly complex because they combine spatial information related to gene organization and temporal relations related to gene and genome evolution. For example, comparison between multiple orthologous and paralogous genomic loci in parallel in a single view is an efficient strategy to rapidly understand the evolutionary history of a locus from a common ancestor. Bioinformatics tools are available to visualize and compare genomes (1–8) but most are restricted to two or three genomes at a time or are stand-alone applications. To represent such complex data in a way that human experts will find useful to accelerate interpretation, Genomicus departs from traditional genome browsers because biological objects (genes, non-coding elements) are not shown to scale but instead are shown schematically. This strategy eliminates intra- and inter-genome variability to focus attention on symbolic representations of shared (homologous) properties.
Here, we present a major extension to the Genomicus database and web interface. Four new clades, including plants, fungi, non-vertebrate metazoa and protists, are now represented in dedicated browsers, bringing the number of eukaryote genomes available for interactive comparisons to 150.
DATA SOURCES AND COMPUTATION
Genomicus displays two kinds of information, the relative position and order of genes in genomes, and the phylogenetic relationships (orthology, paralogy) between genes, extracted from the Ensembl (9) and EnsemblGenomes (10) databases. The first Genomicus server was dedicated to vertebrate genomes (including five non-vertebrate out-groups), based on Ensembl data (11). We now extend the database to other clades, based on EnsemblGenomes data for plants, fungi, non-vertebrate metazoa and protists. Ancestral genome reconstructions are currently available for vertebrates only, but ancestral gene contents are inferred from gene phylogenies for all phyla.
Vertebrates
Synteny and ancestral gene content information
The Genomicus vertebrate server (http://www.dyogen.ens.fr/genomicus) is available since 2010 (11). It is synchronized with Ensembl releases, with an ∼2-week time lag, and all versions are archived online. This server is intensively used by laboratories worldwide and is directly available from the Ensembl treeview display. It provides comparative genomic information on 54 vertebrate genomes and 5 out-groups and extensive information on reconstructed ancestral genomic organization at 49 ancestral nodes. The order of genes is a direct unedited reflection of gene coordinates in Ensembl. Phylogenetic relationships between genes (gene trees) on the other hand are downloaded from Ensembl and edited as follows. Ensembl computes gene trees using the Treebest method, which includes reconciliation with the species tree. At this step, duplication nodes are often inserted in the tree to accommodate branches that are not directly compatible with the species tree (12). This especially happens with ancestral taxa preceding a quick radiation (like placental mammals and percomorph fish). In those taxa, 14.9% of the nodes are flagged as ‘dubious’ by Ensembl (which means they are poorly supported duplications), whereas only 2.4% of the nodes of other taxa are flagged. In these cases, it is often more parsimonious to modify the tree by turning the poorly supported duplication nodes into speciations and pushing the few duplicated genes towards more recent nodes. To achieve this, we use the ‘consistency score’ provided with Ensembl trees, which simply expresses the ratio between the number of species under a duplication node and the number of species with duplicated genes. If the score is 1.0, all species under the duplication node possess a duplicated gene, and the node is maximally supported. We find that duplication nodes with a consistency score <0.33 are generally unreliable and are thus edited as described earlier in the text.
The ancestral gene content and order is computed with AGORA (Algorithms for Gene Order Reconstruction in Ancestors) (13). A full description of the AGORA method and its validation will be published separately. Briefly, gene orders are compared between all possible pairs of genomes to identify adjacent orthologous genes (AOGs). For example, genes a1 and b1 in Species 1 are neighbours, and their respective orthologues a2 and b2 in Species 2 are also neighbours. Under a parsimonious model, such adjacency may be the result of evolutionary conservation, that is, all the ancestral genomes between the two species in the species tree already possessed the adjacency. Alternatively, but much less likely, it may be the result of a fortuitous genomic rearrangement. For a given ancestral genome, AGORA builds a weighted graph where vertices are genes, and edges are created for each AOG. A weight reflects the number of times a given AOG is observed in pairwise comparisons. The graph is then linearized by maximizing weights when vertices are of degrees >2. The linear order of genes (vertices) is considered the most likely ancestral order. AGORA only considers AOG when the transcription orientation is preserved. In release 68 of Genomicus, AGORA processed 19 940 trees comprising 1 050 481 genes in 59 species. After 1711 pairwise genome comparisons, 890 477 ancestral genes were inferred in 49 ancestral genomes. Ancestral gene content information and gene order reconstructions are available on the Genomicus ftp server.
Conserved non-coding elements
Conserved non-coding elements (CNEs) often pinpoint enhancers that control the expression of nearby genes. Visualizing their relative positions to neighbouring genes in multiple species may thus be an additional and important guide to identify the target gene(s). CNE positions can be explored in Genomicus at different levels of sequence conservation. CNEs are defined based on their conservation to human sequences in a range of vertebrate species, using an algorithm that scans the UCSC 46 species multiZ alignment (14) and looks for conserved regions of a minimal length (10 bp) and identity (90% in the 10-bp seed region, further extended by accepting up to three non-conserved columns on each side). This algorithm does not ask for the presence of a fixed set of key species in the alignment, but instead, it only requires that at least eight species be aligned to human. Moreover, it allows substitutions to occur, under a given threshold, in each column of the alignment; a column is considered as ‘conserved’ if at least 88% of its nucleotides are identical. The CNEs are filtered on a minimal size of 20 bp, and we distinguish four levels of conservation with respect to human. They must be conserved in Boreoeutheria genomes (which must include mouse, dog and cow), Mammalians (Boreoeutheria CNEs also conserved in opossum), Amniotes (Boreoeutheria CNEs also conserved in chicken) and Vertebrates (Boreoeutheria CNEs also conserved in at least one among zebrafish, tetraodon, medaka and stickleback). CNEs are excluded from regions overlapping repeated sequences in human only, and from regions overlapping protein coding sequences in all of the species considered. The consensus sequence and the conservation displayed are computed on the 46 species.
In the version of the server based on Ensembl 68 (September 2012), >1.2 million CNEs have been defined, stored in the database and can be explored in the Genomicus PhyloView module (Figure 1). Users can choose to display or hide the CNEs with a tick box in the top-level menu of the page. Hiding CNEs may substantially improve the server and client response. CNEs are displayed between genes in different colours according to the conservation level (green for boreoeutherians, orange for mammals, red for amniotes and blue for vertebrates). A mouse-over on a CNE will highlight orthologous CNEs in other species that lie in the chromosome region displayed. CNEs are represented in two distinct groups within the intergenic space; intronic CNEs are shown in a group abutting the right hand side of their host gene, and intergenic CNEs are evenly distributed in the remaining space. Information on CNEs can be accessed through the top-level panel, such as the corresponding sequences in a multiple alignment, and links to Ensembl and UCSC browsers.
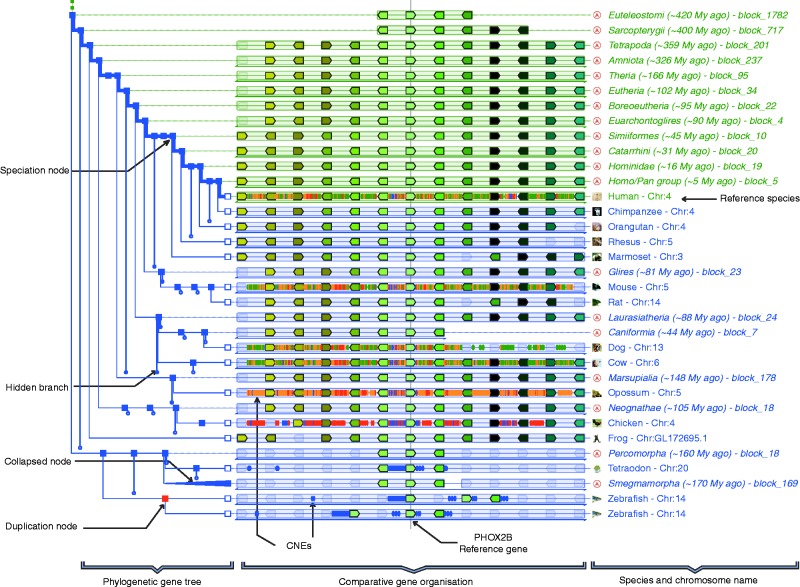
Figure 1.
PhyloView of the paired-like homeobox 2b (PHOX2B) gene in human, the reference gene and reference species for this display, rooted at the Euteleostomi ancestor. This region is remarkably well conserved in the descendants of the ancestral Euteleostomi. PHOX2B is surrounded by a high density of conserved CNEs identified between human and several mammals (green and orange symbols), but also with chicken (red symbols) and fish (blue symbols). To obtain this specific display, the user should (i) query the Genomicus vertebrate server with the human PHOX2B gene; (ii) in the resulting PhyloView display, select ‘View’ and ‘change root species’ to restrict the display to Euteleostomi (89 homologues); (iii) zoom in once to obtain only six genes on each side of the reference gene; (iv) tick the CNE track box in the ‘View’ menu; and (v) hide (e.g. elephant) or collapse (e.g. smegmamorpha) specific species or phylum using the red cross that appears by mouse-over on the corresponding node (to hide) or by directly clicking on the corresponding node (to collapse).
Plants, fungi, metazoa and protists
As an extension to the vertebrate-centred Ensembl, EnsemblGenomes now provides similar data for plant, fungi, protists and non-vertebrate metazoa in exactly the same software environment (10). It, therefore, becomes relatively straightforward to provide additional Genomicus servers to accommodate these new resources, which can be accessed from the following URLs:
Ancestral genome reconstruction will progressively be made available for these clades, starting with plants. The current versions enable evolutionary analysis and exploration, and to study gene expansion or loss, in specific species or groups (Figures 2 and 4). GenomicusPlants displays syntenic information for 19 species, including green algae, monocots, dicots and five eukaryote out-groups (human, Caenorhabditis elegans, yeast, Drosophila and Ciona). GenomicusProtists shows information for 19 extant species, GenomicusFungi for 30 and GenomicusMetazoa for 37. The four new Genomicus databases reflect the organization of the EnsemblGenome databases. Of note, some species within a clade are evolutionarily distant, and it may not make sense to seek any conservation in gene order between some species, especially in groups, such as protists, that are not monophyletic (15). For example, Alveolata (e.g. Plasmodium falciparum) and Amobeozoa (e.g. Dictostelium discoidum) are grouped in GenomicusProtists but share no measurable conservation of gene synteny. Nevertheless, this organization is convenient because users may select specific subgroups based on gene phylogenies to focus on their evolutionary range of interest and use the extended range of species available to rapidly access orthologous genes in different clades.
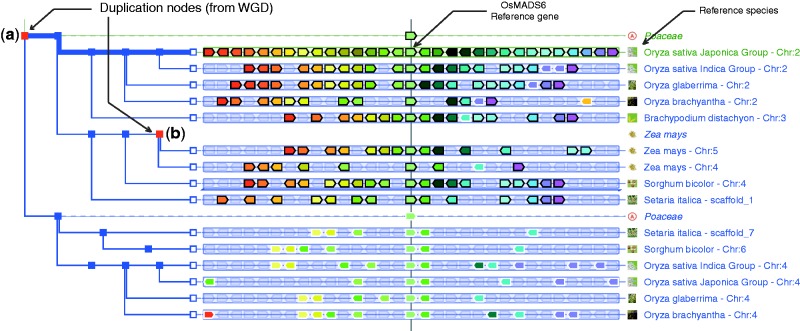
Figure 2.
PhyloView example of a gene in the rice genome (Oryza sativa japonica). The gene OsMADS6 (LOC_Os02g45770) and its orthologues and paralogues are centred on the thin vertical line. The top red square node (a) shows that a duplication of the ancestral OsMAD6 gene took place in the ancestral Poaceae genome. The rice paralogue of OsMAD6 is on chromosome 4 (in green but surrounded with a white line), and the display shows that all other cereals also contain two copies of this gene. Most strikingly, many genes that are neighbours of OsMAD6 also have paralogues in the neighbourhood of its paralogue, and this is true in all cereal genomes. One can, therefore, conclude that this situation is reminiscent of a large segmental duplication, or in this case, of the whole genome duplication (WGD) that took place in the ancestor of cereals. The second red square node (b) shows that a duplication took place in the Zea mays genome. The environment of the reference gene in maize also shows the more recent WGD specific to the Z. mays genome. To obtain this specific display, the user should (i) query the GenomicusPlant server with the LOC_Os02g45770 gene in the rice O. sativa Japonica; (ii) in the resulting PhyloView display, select ‘View’ and ‘change root species’ to restrict the display to the Poaceae root ancestor [28 homologues (dupl. node)]; (iii) select ‘Hide all ancestral nodes’ in the ‘Tree’ menu to hide all ancestors in a single action; and (iv) specifically reveal the Poaceae speciation node and the Z. mays duplication node by clicking on the green cross that appears by a mouse-over on the corresponding nodes.
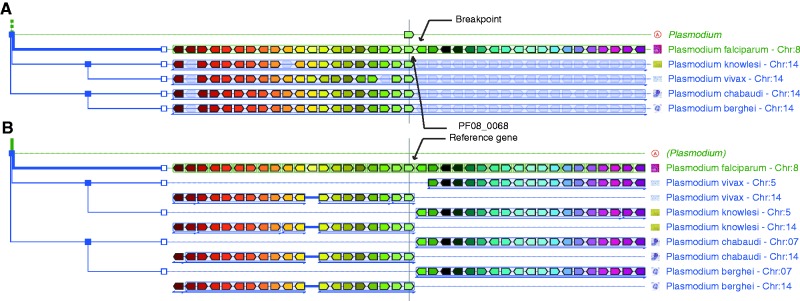
Figure 4.
A chromosome breakpoint in Plasmodium. In Phyloview (A), the reference gene (PF08_0068) in P. falciparum is on the edge of an evolutionary breakpoint, as the gene collinearity stops abruptly when compared with other Plasmodium species. This is confirmed by the AlignView display (B) showing that the synteny is well conserved through Plasmodium but on two different chromosomes for the four other Plasmodium. To obtain display shown in A, the user should (i) query the GenomicusProtists server with the PF08_0068 gene in P. falciparum; (ii) select ‘View’ and ‘change root species’ to restrict the display to Plasmodium species (eight homologues); (iii) zoom out; (iv) adjust the zoom by clicking four times on the ‘-’ sign in the top frame so that four more genes are shown on either sides of the reference gene; and (v) hide the two ancestral Plasmodium lines by clicking on the red cross that appears by mouse-over on the corresponding nodes. The user can then switch to the AlignView display by selecting the relevant option in the ‘View’ menu.
The possibility of directly interpreting the conservation of gene order between species, as presented in the Genomicus databases, depends on the quality of the underlying genomic data. Perfectly contiguous genome assemblies and exhaustive protein coding gene annotations do not exist for any genome yet. The most complete data set concern a few model organisms, such as Saccharomyces cerevisiae, C. elegans, Drosophila melanogaster, Arabidopsis thaliana, Homo sapiens, Mus musculus or Danio rerio. For many others, assemblies are increasingly based on whole genome shotgun sequencing using short-read technologies, leading to fragmented chromosomes assemblies. Protein-coding gene annotations depend on the contiguity of these assemblies and on additional resources (e.g. expression data) that are not always available, leading to partial gene structure and gene content annotations. Together, these limits bear on the quality of the phylogenetic reconstructions and on orthology and paralogy assignations. GenomicusVertebrates provides an option to automatically collapse low coverage genome assemblies to clear the display of such data that generally provides little additional information.
GENOMICUS VIEWS
The home page of each Genomicus server invites the user to enter a gene of interest, which will be defined as ‘reference gene’ and belongs to a ‘reference species’. This ‘reference gene’ is the starting point to explore its genomic context and its evolution. The default view (PhyloView) can be accessed by a gene name (ex: Phox2b) or an Ensembl geneID (ex: ENSDARG00000024771).
PhyloView
The PhyloView page (Figures 1 and 2) shows the order of genes in the neighbourhood of the reference genes and the order of their orthologues and paralogues in different species. Species are shown only if they contain an orthologue or a paralogue of the reference gene. In this view, the reference gene and its homologues are displayed over a vertical central line, in green. Neighbouring genes in the reference species are displayed with different colours, and each gene will share its colour with its homologues in other species. The species are organized according to the phylogenetic gene tree of the reference gene, drawn on the left. In this tree, blue and red nodes represent a speciation and a duplication, respectively. In the default view, information on low coverage species is hidden (defined by a branch ending with a little blue circle). Hidden branches can be expanded by a simple click.
AlignView
The AlignView page shows an alignment between genes contained within the genomic region of the reference gene and all their respective orthologues in other species. Unlike PhyloView, AlignView represents the genomic environment in all species that have, at least, two collinear orthologous genes with the genomic region of reference. A species can thus be represented even if it does not possess any orthologues of the reference gene. The species are organized according to the species tree, drawn in blue on the left size of the window. The genes of a given species can be spread over multiple lines if the reference region is distributed over several chromosomes. This view allows an intuitive visualization of gene loss or gain during evolution (Figure 3), and confirmation of potential breakpoint (Figure 4).
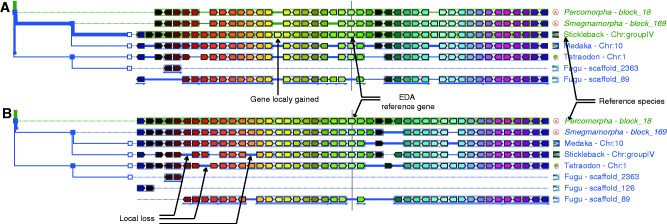
Figure 3.
AlignView to analyse the neighbourhood and local synteny around the Ectodysplasin A (EDA) gene. (A) When the reference species is stickleback, other species, such as medaka and tetraodon, possess all orthologous copies from this locus on a single chromosome, whereas the orthologues are distributed over two scaffolds in Fugu. It is also clear that stickleback has one extra gene (in yellow) that is absent in the three other fish and all ancestors, perhaps, therefore, resulting from a specific gain in this species. (B) When the reconstructed ancestral Percomorpha genome is used as reference, local gene losses in different descendent branches become apparent. To obtain the displays shown in A, the user should (i) query the GenomicusVertebrate server with the EDA gene in stickleback; (ii) select ‘View’ and ‘change root species’ to restrict the display to Percomorpha (eight species) in the AlignView column; (iii) zoom out once and shift the display by one gene to the left using the relevant buttons in the top frame; and (iv) hide Tilapia to simplify the display. The view shown in B is then obtained after selecting the Percomorpha ancestral EDA gene and clicking on ‘switch to this gene as reference’ in the top frame.
Top menu features
Information on element in the display
The top-level menu provides information on the default reference gene, or on the selected gene in the display. It allows switching to a new reference gene or species and provides links to external websites (Ensembl, EnsemblGenome, UCSC, NCBI) that open in a new window. The same kind of information can be accessed when selecting a CNE.
Browsing in the reference region
User can restrict or extend the default display region to 3–40 genes and browse the upstream or downstream region of the reference gene with the zoom and arrow menu.
Interactive specification of output graphics
In the views described earlier in the text, the user can select the species of interest and can hide every node or groups of species that are considered to be non-informative. An ancestral node and all the descendant branches can be collapsed in one click and replaced by a triangle in the tree or can be completely hidden by clicking on the red cross that appears on mouse-over (for ancestral and modern species). Clicking on the green cross or the triangle expands collapsed or hidden branches, respectively. This is particularly useful to streamline the display before printing or exporting, so that only the species of interest are shown.
Focus on paralogues
Users interested in comparing the organization of genes between paralogous loci may do so in one click. After selecting the reference gene, selecting this option will automatically collapse and hide all the branches except the branches leading to the paralogues of the gene of interest.
Hide all ancestral species, hide all out-groups of specific species
To avoid numerous actions on the graphical part of the display, the top menu allows hiding all information relative to ancestral gene content in one action, or hiding all out-groups of one ancestral node in one action.
Exporting data and images
Each graphical view can be exported. Three options are available. A text-based description of the data summarizes the information for each node and each modern species, with gene names and their relative position. This may be useful for further automatic processing using ad hoc script. The information may also be exported in Scalable Vector Graphics (SVG) format, as displayed or with all nodes being expanded. The exported SVG format can easily be imported in Inkscape, for example, if additional work editing is required. Alternatively, depending on the browser, the page may be printed as a PDF file.
GENOMICUS SOFTWARE IMPLEMENTATION
All the data in Genomicus are stored in MySQL databases. The interface is composed of Perl scripts and modules. It runs on an Apache2 server, using mod_perl. The different modules like AlignView and PhyloView generate pages embedded with inline-SVG drawings in XHTML. Javascript is used only for the information panel and mouse-over information and is retrieved with Asynchronous JavaScript and XML (AJAX) calls. The Genomicus browser is currently optimized for Firefox. It runs on Chrome and Safari and can be used with Internet Explorer if the Google Chrome Frame plugin is installed.
Genomicus sources and MySQL schema are available on request.
FUTURE PLANS
The Genomicus database provides a simple and intuitive approach to compare extant and ancestral genomes in a local gene environment. Ongoing methodological development focuses on a more global view of the comparison. For example, we plan to enable karyotype comparison and dotplot matrices linked to PhyloView and AlignView. Additional information will also be added in the two main displays, such as a colour gradient reflecting the sequence similarity between orthologues and paralogues and dN/dS information. Future developments will also address the possibility to perform a Basic Local Alignment Search Tool (BLAST) comparison with the genomes.
FUNDING
Centre National de la Recherche Scientifique (CNRS); Agence Nationale de la Recherche [Ancestrome Project ANR-10-BINF-01-03, ANR Blanc-PAGE ANR-2011-BSV6-00801]; 7th framework programme of the European Union [NeuroXsys Project HEALTH-F4-2009-223262]. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Pierre Vincens for assistance with computer systems administration, numerous users for feedback on the Genomicus interface and the Ensembl and EnsemblGenome projects for providing integrated comparative genomic data to the community.
REFERENCES
- 1.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan X, Stein L, Brendel V. SynBrowse: a synteny browser for comparative sequence analysis. Bioinformatics. 2005;21:3461–3468. doi: 10.1093/bioinformatics/bti555. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Su Y, Mackey AJ, Kraemer ET, Kissinger JC. SynView: a GBrowse-compatible approach to visualizing comparative genome data. Bioinformatics. 2006;22:2308–2309. doi: 10.1093/bioinformatics/btl389. [DOI] [PubMed] [Google Scholar]
- 4.Courcelle E, Beausse Y, Letort S, Stahl O, Fremez R, Ngom-Bru C, Gouzy J, Faraut T. Narcisse: a mirror view of conserved syntenies. Nucleic Acids Res. 2008;36:D485–D490. doi: 10.1093/nar/gkm805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López MD, Samuelsson T. eGOB: eukaryotic Gene Order Browser. Bioinformatics. 2011;27:1150–1151. doi: 10.1093/bioinformatics/btr075. [DOI] [PubMed] [Google Scholar]
- 6.Revanna KV, Chiu CC, Bierschank E, Dong Q. GSV: a web-based genome synteny viewer for customized data. BMC Bioinformatics. 2011;12:316. doi: 10.1186/1471-2105-12-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soderlund C, Bomhoff M, Nelson WM. SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011;39:e68. doi: 10.1093/nar/gkr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, Humphrey JC, Hughes DST, Keenan S, Kerhornou A, Koscielny G, et al. Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–D97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muffato M, Louis A, Poisnel CE, Roest Crollius H. Genomicus: a database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics. 2010;26:1119–1121. doi: 10.1093/bioinformatics/btq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muffato M. 2010. Reconstruction de génomes ancestraux chez les vertébrés. Ph.D. Thesis. Université d'Evry Val d'Essonne. [Google Scholar]
- 14.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol. Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]