Abstract
We report here the construction of engineered endonuclease database (EENdb) (http://eendb.zfgenetics.org/), a searchable database and knowledge base for customizable engineered endonucleases (EENs), including zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). EENs are artificial nucleases designed to target and cleave specific DNA sequences. EENs have been shown to be a very useful genetic tool for targeted genome modification and have shown great potentials in the applications in basic research, clinical therapies and agricultural utilities, and they are specifically essential for reverse genetics research in species where no other gene targeting techniques are available. EENdb contains over 700 records of all the reported ZFNs and TALENs and related information, such as their target sequences, the peptide components [zinc finger protein-/transcription activator-like effector (TALE)-binding domains, FokI variants and linker peptide/framework], the efficiency and specificity of their activities. The database also lists EEN engineering tools and resources as well as information about forms and types of EENs, EEN screening and construction methods, detection methods for targeting efficiency and many other utilities. The aim of EENdb is to represent a central hub for EEN information and an integrated solution for EEN engineering. These studies may help to extract in-depth properties and common rules regarding ZFN or TALEN efficiency through comparison of the known ZFNs or TALENs.
INTRODUCTION
Engineered endonucleases (EENs) are designed to bind and cleave specific DNA sequences in vitro or in vivo. Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are the two most widely used customizable types of EENs that exhibit many features in common (1). They recognize target sequences via engineerable DNA-binding domains and cut the DNA by non-specific nuclease domains (2,3). With these artificial endonucleases, site-specific DNA double-stranded breaks (DSBs) can be theoretically induced in any given genome, by which targeted genome modification could be easily achieved. Various genetic manipulations, such as gene disruption, gene correction and gene addition, have been designed and achieved in many organisms as well as in cultured cells based on EEN techniques. It is specifically essential for gene targeting in those species where no other reverse genetics approaches are available (4–8). Other types of EENs are also reported, such as triplex-forming oligonucleotides (9–12), engineered homing endonucleases (HEs) (13–15) and fusion proteins of the DNA-binding domains of zinc fingers or HE with full-length restriction endonucleases (16,17); however, they are much more difficult to customize than ZFNs and TALENs, which are based on a modular-repeat structure. Furthermore, a database called LAHEDES for HE-engineering has been reported (18).
Most ZFNs and TALENs function as dimers. Each monomer of these EENs consists of an artificially constructed hybrid protein containing a specific DNA-binding domain, which is derived from zinc finger proteins (ZFPs) for ZFNs (19), or transcription activator-like (TAL) effectors (TALEs) for TALENs (20) and a non-specific cleavage domain, which usually comes from the FokI restriction endonuclease (2) as well as a linker peptide between these two domains and other framework sequences. The DNA-binding domain of ZFPs or TALEs consists of an array of tandem repeat units, each of which recognizes and binds to one or more nucleotide targets in one strand of the DNA. As for ZFP, each zinc finger repeat approximately targets 3 nt (called a triplet), but internal and external context-dependent effects of neighboring fingers can influence the efficiency and specificity of the corresponding target-protein relations (21). No accurate relationship has been discovered and several strategies have been developed to screen for and construct efficient and specific ZFPs and ZFNs. In contrast, TALE domains show a more predictable one-to-one correspondence between repeat units and their single-nucleotide targets, and no screening is required for TALEN engineering (22,23). Although hundreds of ZFNs or TALENs have been constructed and tested, the targeting efficiency of these EENs varies significantly. In fact, the efficiencies of TALENs in the same work or even targeting the same locus may vary from low to high, or even showing no targeting activity (24–27). Unfortunately, now it is still difficult to predict the efficiency of a given ZFN of TALEN, largely due to the lacking of sufficient knowledge about the properties of these EENs. Through analyzing the rapidly expanding information of these EENs from various resources, it may help to extract common rules regarding the efficiencies of TALENs or ZFNs through comparison of all the evaluated TALENs or ZFNs, respectively. On the other hand, the specificity and potential toxicity of TALENs still need further and careful investigation. In certain applications, the technology of ZFN is more mature, e.g. ZFNs are being tested in gene therapy trials (21,28–30), and are reported recently to be able to be delivered into cultured cells in the form of purified proteins directly (31). The properties of these two types of EENs have been compared with each other in some of the recent works (26,27,32,33).
Other factors in the customizable EEN engineering for TALENs and ZFNs, such as variants of FokI cleavage domains (34–40), as well as the relationship between the length of linker peptides and the length of spacer between the two EEN monomer binding sites (41–46), can also affect the efficiency and specificity of these EENs. Methodologies for the customizable EEN construction and genome modification have been optimized for best performance. The efficiency of EENs and the detection methods as well as the specificity evaluated by off-target tests have been reported previously (47,48). Collection and comparison of the corresponding information may be useful for reviewing/reusing existing EENs and engineering new TALENs or ZFNs.
Some databases such as ZifBASE (49) and ZiFDB (50) have been developed to collect the information of some of the ZFP domains but not the nucleases themselves. So far, hundreds of efficient ZFNs and TALENs have been reported and many methods for their engineering and construction including high-throughput ones (24,51) have been developed. However, a database systematically collecting and classifying the information of all the currently known customized EENs and their targets is unfortunately lacking.
Some tools are available to predict/search for candidate EEN target sites in given DNA sequences. TALE-NT (52), ZiFiT (53) and idTALE (54) are predominant tools for TALEN/TALE target design. ZiFiT also supports ZFN target design and selection covering various strategies, whereas ZFN Sequence Tag and other web-tools (55–57) only support particular types of ZFN design. On the other hand, several strategies have been developed to screen for and construct customized EENs. Polymerase chain reaction (PCR)-based Golden Gate cloning (58,59), vector-based Golden Gate cloning (60–64), Unit Assembly (65), REAL/REAL-Fast (33), dTALE-assembly (54), FLASH (24) and ICA (51) are designed to construct the long and highly repetitive TALE DNA-binding domains, which are difficult to synthesize by ordinary PCR or modular cloning. Modular Assembly (55,66,67), OPEN (68), CoDA (69) and other public (70,71) or commercial strategies (72) are developed to screen for and obtain ZFP domains showing high target binding activity. Apart from these methods, several platforms such as Addgene (http://www.addgene.com) offer plasmids or kits and protocols for EEN engineering with several selections of different methods or strategies, which might be difficult to find and compare all the existing resources and make a proper decision if one does not work on and is not familiar with the field of EEN engineering. A knowledge base providing a route with simple descriptions for and links to these tools and resources will allow users to take it as a starting point and find the most appropriate one for themselves.
Here, we report the construction of EENdb, the first database and knowledge base dedicated to a complete and detailed collection of all the currently available and evaluated ZFN and TALEN data extracted and curated from publications and other sources. It also integrates different EEN (ZFN and TALEN) engineering resources, to provide a relatively complete solution for searching for known EENs as well as for design and construction of new customized ZFNs and TALENs. In current version of EENdb and here below, EENs refer to ZFNs and TALENs.
MATERIALS AND METHODS
Data sources
We manually extracted the information of all ZFNs and TALENs reported to be effective, either targeting endogenous loci or designed for artificial sequences, directly from published research articles, or reconstructed the necessary information if it was not given directly. Non-functional TALENs and ZFNs were also included for comparison and analysis, but numerous inactive ZFNs from screening attempts were omitted. The data of some TALENs showing no detectable activity were also collected via direct submission by the authors.
In total, more than 400 records of ZFNs, more than 300 records of published TALENs and 24 records of directly submitted TALENs have been collected and curated so far. Redundant EENs from the same or different sources were merged. The core information in the database consists of targeting site sequences, types of ZFN and TALEN forms, critical peptide sequence of the DNA-binding domains, linker peptides and frameworks, FokI variants and other alternate cleavage domains, ZFP or TALE construction strategies and genome modification methods. EEN efficiency with the detection methods and EEN specificity with off-target sites tested were also included if available. In the whole target site for a pair of EENs, the ZFP- or TALE-binding sequences (i.e. half-sites) and spacer sequences between the two half-sites were carefully differentiated.
An NIH-sponsored project aiming to target endogenous genes in zebrafish (Danio rerio) regularly constructs arrays of ZFNs and TALENs with only ZFP- or TALE-binding activities tested and releases them in Addgene (http://www.addgene.com/zfc/arrays/ and http://www.addgene.com/talengineering/TALENzebrafish/). EENdb has not collected the information of these EENs unless the cleavage activities were tested and reported. However, a prompt message appears and indicates to the project web pages when users list EENs of all species or of zebrafish in EENdb.
To complement with the EEN tables, we also collected the information of natural and artificial ZFPs (only the C2H2 type, which is used as the framework for engineered ZFPs), including those with only binding capability but not cleavage activity reported and artificial TALEs. The 7-aa variable region of each finger of ZFPs (including ZFNs) and the repeat-variable di-residue (RVD) of each repeat unit of TALEs, which are considered to be the most critical elements determining DNA-binding specificity, were extracted and displayed in association with their target nucleotides.
Design and implementation
EENdb is implemented with open source technologies. The data are stored in a MySQL relational database. The web site is written in PHP scripts and the service is provided by an Apache/PHP server.
RESULTS
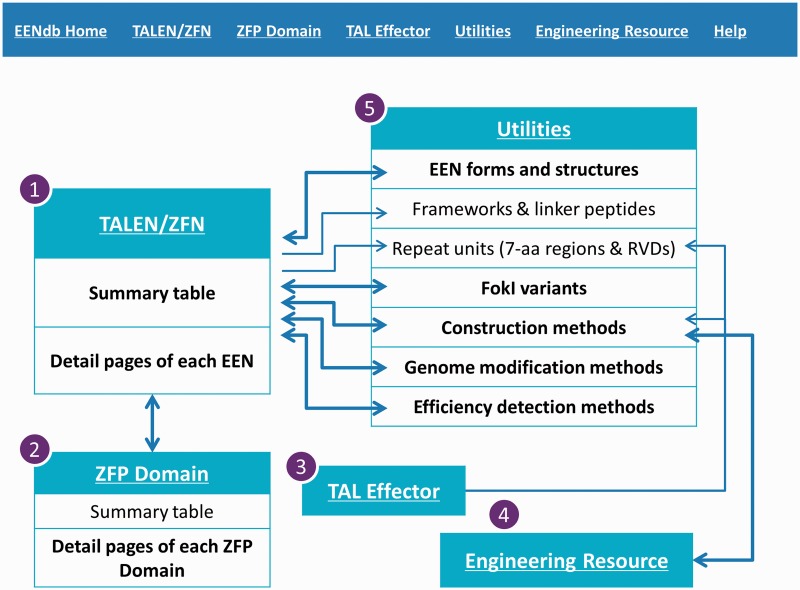
EENdb consists of five interrelated sections (Figure 1). The main section is the collection and summary of the information about EENs (including TALENs and ZFNs) called ‘TALEN/ZFN’. ‘ZFP Domain’ and ‘TAL Effector’ are two sections built to complement with the main section, whereas ‘ZFP Domain’ provides the collection of the zinc finger DNA-binding domains, especially the 7-aa regions from ZFNs and other ZFPs; ‘TAL Effector’ collects the information of the TALE DNA-binding domains from TALE proteins other than TALENs. Details about the components and information of EENs, such as variants of the FokI cleavage domains, the scaffolds/frameworks and linker peptides between the DNA-binding domains and the cleavage domains, as well as the information about the construction and application of EENs, are summarized in the section of ‘Utilities’ and linked to other EEN datasets in each corresponding field. The last section named ‘Engineering Resource’ provides short descriptions of and links to all the available external resources for EEN engineering and is also linked to the EEN construction methods summarized in the ‘Utilities’.
Figure 1.
The menu banner of EENdb and the relationship between the five main sections of the database. Contents and organization of these sections are described in the text. The arrows represent links between the sections and web pages.
The dataset of EENs
Collection of the essential information about EENs is the major component of EENdb. Users can list all TALENs and/or ZFNs designed to target a special species, or easily search by gene symbols, gene IDs, whole or partial target sequences (degenerate nucleotides are also supported), or reference PMIDs (PubMed IDs) or surnames of the first authors from the ‘TALEN/ZFN’ page (Figure 2A). Results of corresponding ZFNs, TALENs or integration of the two types of EENs are returned, according to the search option set by users. In the search result table or the table listed by species, summaries of EEN information are listed (Figure 2B). More details can be accessed through the detail page by clicking on the EENdb IDs in the table.
Figure 2.
Screenshots of EENdb. (A) The section of TALEN/ZFN. (B) Part of the summary table of listing all EENs in zebrafish. (C) Part of the web page of FokI variants in the ‘Utilities’ section.
For each EEN, the following information is displayed in the detail page; some of the information is omitted or shown in abbreviation in the summary table.
EENdb ID
EEN data are collected and classified according to their target sites. ID of a ZFN record usually has the format ‘ZNxxxx’ or ‘ZNAxxx’ and ID of a TALEN has the format ‘TNxxxx’ or ‘TNAxxx’, in which an ‘x’ refers to a digit. ‘ZNxxxx’ and ‘TNxxxx’ are assigned to EENs targeting natural sites, whereas ‘ZNAxxx’ and ‘TNAxxx’ are for EENs targeting artificially synthesized DNA sequences. Groups of different EENs targeting similar sequences from the same or different literature are given serial IDs with one-letter suffixes. For example, TN0002, TN0002B, TN0002C and TN0002D all target the same or overlapped site of human CCR5 gene. An ‘-Txxx’ suffix indicates that this record contains a previously reported EEN (i.e. the same EEN) but targeting another DNA sequence. In most cases, it represents an off-target site. For example, TN0031-T002 targets CCR2, an off-target gene of TN0031, which is originally designed to target CCR5 gene. In summary tables of the list of a particular species or search results, the EENs are sorted as a default by publication PMID (approximately in the order of publication date) so that EENs from the same publication are arranged together. Alternatively, it can be changed to group by EENdb ID, which is also used in the EEN detail pages, thereby related records with same target sequence and/or off-targets of same EENs are centralized for comparison.
EEN type or form
Whether an EEN is a ZFN or a TALEN can be easily distinguished from the first letter of the ID. In addition, most of the EENs are functional in dimers and the FokI domains are usually fused in the C-terminus of the monomers, but exceptions do exist (73–78). A description of rarely used EEN form and a link to a page with detailed explanation in the ‘Utilities’ section is provided under the corresponding EENdb ID.
Target site sequence
The sequence of the whole target site recognized by EEN pairs is carefully extracted from or constructed based on publications. The half-sites (i.e. the binding site for a single EEN) of the whole target site are shown in uppercase; the strands bound by the ZFP- or TALE-binding domains are underlined. The spacers are revealed as lowercase letters. Additional one base pair outside the half-sites is also provided for it is important for ZFNs and TALENs in some cases, e.g. to distinguish whether the additional nucleotide is the most commonly used nucleotide T for TALENs (3,24), or to consider the context-dependent effect of ZFN fingers (79,80). The target sequences can be searched by either the forward or the reverse strands.
Other information of the target site
For natural targets, the English and Latin names of species and the gene or genomic locus represented by Ensembl IDs, Ensembl Genomes IDs or RefSeq Accessions are given. Length of the spacer, numbers of fingers in a ZFN and lengths of half-sites of a TALEN are calculated and displayed.
DNA-binding domains of EEN monomers
The key amino acids of the DNA-binding domains are shown here. For ZFNs, the 7-aa variable regions of each finger and a link to the ‘ZFP Domain’ section are provided. For TALENs, four most commonly used RVDs, each recognizing its own corresponding single-nucleotide target (i.e. NI for nucleotide A, HD for C, NG for T and NN for G), are considered as ‘standard code’ of RVDs (22,23); other non-standard ‘alternative code’ of RVDs (e.g. NK or NH for G, NG for mC or 5-methylcytosine) (20,81–83) and off-targeted RVDs are marked with colors different from the ‘standard’ ones for identification.
Other components of the EEN protein
The types of linker peptides between the DNA-binding domains and FokI domains, the variants of FokI cleavage domains as well as the screening and construction methods or strategies for the DNA-binding domains are included if they are known. These items are linked to the related pages in the section of ‘Utilities’.
Effectiveness of EEN
Each EEN record also contains the modification method of the target locus [e.g. non-homologous end joining (NHEJ) or homologous recombination (HR)], efficiency of the EENs and the detection methods and in vitro cultured cell lines tested if available, heritability of EEN-induced mutations at organism level and specificity of the EEN revealed by the result of off-target evaluation. The modification and efficiency detection methods are also linked to the corresponding ‘Utilities’ pages for detailed explanation. In some cases, comparisons or collaborations with other EENs are indicated as comments.
References
PMID with a link to the first publication reporting certain pair of EENs is provided, or other information is listed for directly submitted records.
User commenting system
In each detail page of the EEN records, a simple commenting system is available; any correction, complementarity and communication can be contributed by users either with this system or through contacting the administrators of EENdb by email.
The dataset of ZFP domain and TALE
Natural or artificial ZFP domains and artificial TALEs compose two other sections of EENdb in parallel to the section of TALEN/ZFN. The dataset of ZFP domains consists of more than 1000 records and provides much more information about ZFP-binding domains for data mining, e.g. discovering the relationship between the key peptide sequences and the target nucleotide sequences of the nucleases based on statistical analysis of known ZFPs. The dataset of TALEs provides a link to a list of natural TALEs and collects TALEs beyond TALENs, which are both rich in ‘alternative code’ or off-targeted RVDs and may be helpful for researchers who are interested in the analysis of TALE-binding activities and discovering more information of the DNA-binding properties of TALEs.
The utilities of EENdb
One of the aims of EENdb is to build a knowledge base about EENs. We invested a great deal of effort to organize and refine the information and utilities of EENdb, which offers a global view of the emerging and fast developing ZFN and TALEN technology and resources. In the ‘Utilities’ section, detailed information of ZFN and TALEN forms and structures, frameworks and linker peptides, FokI variants and other alternate cleavage domains, repeat units of DNA-binding domains, ZFP- or TALE-construction strategies, genome modification methods and efficiency detection methods are provided (Figure 1). These data can help users to retrieve details about EENs, compare and choose optimal parameters, etc. Most of the utilities also provide links to the ‘TALEN/ZFN’ section to filter all EENs matching defined conditions.
For example, the page of FokI variants (Figure 2C) lists all known FokI pairs that can only work in hetero-dimers to reduce non-specificity, or that can enhance catalytic activity, or that participate in building a nickase (only cleave one strand of the target DNA) rather than a nuclease (37–39). Users can list the records of all reported EENs containing specific pairs of FokI domains from the EEN dataset via the links under each FokI variants. The sequences of variants with the mutations highlighted can also be found in this page.
Other examples for the usage of the ‘Utilities’ section are the pages of repeat units of DNA-binding domains, genome modification methods and efficiency detection methods. Similar to the page for FokI variants, users can list the records of all EENs reported to induce HR in different organisms, or all TALENs with particular non-standard ‘alternative’ RVD via corresponding links. This information may help researchers to choose appropriate method for their own experiments. Only as an example of this attempt, one can easily conclude from the list of all the EENs tested in zebrafish that NHEJ is the only type of genome modification induced by EENs in this species except for one very recent report (84), and the most frequently used detection methods in this species are restriction enzyme-resistance assay (after PCR) and direct sequencing.
EEN engineering tools and resources
As described above, many EEN target prediction tools and construction resources are available for the public. The ‘Engineering Resource’ section of EENdb generates links and provides short descriptions and comments of these tools and resources, including candidate TALEN/TALE target site prediction tools, candidate ZFN/ZFP target site prediction tools, target- or off-target-site finder of given EENs, resources of EEN construction protocols and materials and other links to news sites and newsgroups. Applicable construction methods supported by any available resources are provided with links to the corresponding pages in the section of ‘Utilities’.
Access
EENdb can be freely accessed via http://eendb.zfgenetics.org/. The data of EENdb can be downloaded as tab-separated version (TSV) plain text format in the ‘Help’ section of the web site.
EENdb welcomes researchers to submit the information of their newly constructed EENs and related experimental data by email, especially the negative data not intended for publication. The feedback form and email address can be found in the ‘Help’ section of the web site.
DISCUSSION
Continuous updates of the EENdb database will be offered. As more and more EENs are expected to be reported, EENdb will expand to offer links to show custom tracking of known EEN sites in Ensembl and UCSC Genome Browser when necessary.
EENdb has collected the references of all publications first reporting a particular ZFN or TALEN. Other references related to EENs, such as new applications of a known EEN, will also be included in future releases of EENdb.
FUNDING
The 973 program [2012CB945101, 2011CBA01000 and 2011CBA01102]; National Natural Science Foundation of China (NSFC) [31110103904 and 30730056]; National Science and Technology Infrastructure Program [2009FY120100]; 111 Project [B06001]. Funding for open access charge: 111 Project [B06001].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank I.C. Bruce for language editing of the manuscript; P. Huang, Z. Wang, X. Tong Y. Zu and D. Liu for helpful discussions; P. Huang, Z. Wang, Y. Shen, W. Liang, Z. Luo, Q. Wu, W. Li and D. Liu for providing the information of some of the non-functional TALENs; J. Luo for the support and discussion on issues regarding bioinformatics and database management; Y. Shen, Y. Gao and J. Zhang for lab management and for the collection, organization and maintenance of the information and materials of the ZFNs and TALENs from our lab.
REFERENCES
- 1.Baker M. Gene-editing nucleases. Nat. Methods. 2012;9:23–26. doi: 10.1038/nmeth.1807. [DOI] [PubMed] [Google Scholar]
- 2.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao A, Hu YY, Wang WY, Yang ZP, Wang ZX, Huang P, Tong XJ, Zhang B, Lin S. Progress in zinc finger nuclease engineering for targeted genome modification. Hereditas. 2011;33:665–683. doi: 10.3724/sp.j.1005.2011.00665. [DOI] [PubMed] [Google Scholar]
- 5.Huang P, Zhu Z, Lin S, Zhang B. Reverse genetic approaches in zebrafish. J. Genet. Genomics. 2012;39:421–433. doi: 10.1016/j.jgg.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeFrancesco L. Move over ZFNs. Nat. Biotechnol. 2011;29:681–684. doi: 10.1038/nbt.1935. [DOI] [PubMed] [Google Scholar]
- 8.McMahon MA, Rahdar M, Porteus M. Gene editing: not just for translation anymore. Nat. Methods. 2012;9:28–31. doi: 10.1038/nmeth.1811. [DOI] [PubMed] [Google Scholar]
- 9.Silanskas A, Zaremba M, Sasnauskas G, Siksnys V. Catalytic activity control of restriction endonuclease–triplex forming oligonucleotide conjugates. Bioconjug. Chem. 2012;23:203–211. doi: 10.1021/bc200480m. [DOI] [PubMed] [Google Scholar]
- 10.Dahmen V, Kriehuber R. Cytotoxic effects and specific gene expression alterations induced by I-125-labeled triplex-forming oligonucleotides. Int. J. Radiat. Biol. 2012;88:972–979. doi: 10.3109/09553002.2012.702298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolganova NA, Shchyolkina AK, Chudinov AV, Zasedatelev AS, Florentiev VL, Timofeev EN. Targeting duplex DNA with chimeric alpha, beta-triplex-forming oligonucleotides. Nucleic Acids Res. 2012;40:8175–8185. doi: 10.1093/nar/gks410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govan JM, Uprety R, Hemphill J, Lively MO, Deiters A. Regulation of transcription through light-activation and light-deactivation of triplex-forming oligonucleotides in mammalian cells. ACS Chem. Biol. 2012;7:1247–1256. doi: 10.1021/cb300161r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinstiver BP, Wolfs JM, Kolaczyk T, Roberts AK, Hu SX, Edgell DR. Monomeric site-specific nucleases for genome editing. Proc. Natl Acad. Sci. USA. 2012;109:8061–8066. doi: 10.1073/pnas.1117984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baxter S, Lambert AR, Kuhar R, Jarjour J, Kulshina N, Parmeggiani F, Danaher P, Gano J, Baker D, Stoddard BL, et al. Engineering domain fusion chimeras from I-OnuI family LAGLIDADG homing endonucleases. Nucleic Acids Res. 2012;40:7985–8000. doi: 10.1093/nar/gks502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulge UY, Baker DA, Monnat RJ., Jr Comprehensive computational design of mCreI homing endonuclease cleavage specificity for genome engineering. Nucleic Acids Res. 2011;39:4330–4339. doi: 10.1093/nar/gkr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schierling B, Dannemann N, Gabsalilow L, Wende W, Cathomen T, Pingoud A. A novel zinc-finger nuclease platform with a sequence-specific cleavage module. Nucleic Acids Res. 2012;40:2623–2638. doi: 10.1093/nar/gkr1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonfara I, Curth U, Pingoud A, Wende W. Creating highly specific nucleases by fusion of active restriction endonucleases and catalytically inactive homing endonucleases. Nucleic Acids Res. 2012;40:847–860. doi: 10.1093/nar/gkr788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor GK, Petrucci LH, Lambert AR, Baxter SK, Jarjour J, Stoddard BL. LAHEDES: the LAGLIDADG homing endonuclease database and engineering server. Nucleic Acids Res. 2012;40:W110–W116. doi: 10.1093/nar/gks365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg JM. Zinc finger domains: hypotheses and current knowledge. Annu. Rev. Biophys. Biophys. Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- 20.Scholze H, Boch J. TAL effector-DNA specificity. Virulence. 2010;1:428–432. doi: 10.4161/viru.1.5.12863. [DOI] [PubMed] [Google Scholar]
- 21.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophy. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 22.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 23.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 24.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips MI. Gene, stem cell, and future therapies for orphan diseases. Clin. Pharmacol. Ther. 2012;92:182–192. doi: 10.1038/clpt.2012.82. [DOI] [PubMed] [Google Scholar]
- 29.Olender S, Wilkin TJ, Taylor BS, Hammer SM. Advances in antiretroviral therapy. Top. Antivir. Med. 2012;20:61–86. [PMC free article] [PubMed] [Google Scholar]
- 30.Overlack N, Goldmann T, Wolfrum U, Nagel-Wolfrum K. Gene repair of an usher syndrome causing mutation by zinc-finger nuclease mediated homologous recombination. Invest. Ophthalmol. Vis. Sci. 2012;53:4140–4146. doi: 10.1167/iovs.12-9812. [DOI] [PubMed] [Google Scholar]
- 31.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Um E, Cho SR, Jung C, Kim JS. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods. 2011;8:941–943. doi: 10.1038/nmeth.1733. [DOI] [PubMed] [Google Scholar]
- 33.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 35.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 36.Guo J, Gaj T, Barbas CF., 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J. Mol. Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 38.Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22:1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, Cathomen T, Scharenberg AM, Joung JK. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, Hua KL, Yan JJ, Babiarz JE, Gregory PD, et al. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 2012;22:1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu Y, Bhakta MS, Segal DJ. Restricted spacer tolerance of a zinc finger nuclease with a six amino acid linker. Bioorg. Med. Chem. Lett. 2009;19:3970–3972. doi: 10.1016/j.bmcl.2009.02.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol. Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kandavelou K, Ramalingam S, London V, Mani M, Wu J, Alexeev V, Civin CI, Chandrasegaran S. Targeted manipulation of mammalian genomes using designed zinc finger nucleases. Biochem. Biophys. Res. Commun. 2009;388:56–61. doi: 10.1016/j.bbrc.2009.07.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isalan M. Zinc-finger nucleases: how to play two good hands. Nat. Methods. 2012;9:32–34. doi: 10.1038/nmeth.1805. [DOI] [PubMed] [Google Scholar]
- 47.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 49.Jayakanthan M, Muthukumaran J, Chandrasekar S, Chawla K, Punetha A, Sundar D. ZifBASE: a database of zinc finger proteins and associated resources. BMC Genomics. 2009;10:421. doi: 10.1186/1471-2164-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu F, Sander JD, Maeder M, Thibodeau-Beganny S, Joung JK, Dobbs D, Miller L, Voytas DF. Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. 2009;37:D279–D283. doi: 10.1093/nar/gkn606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Piatek MJ, Atef A, Piatek A, Wibowo A, Fang X, Sabir JS, Zhu JK, Mahfouz MM. Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification. Plant Mol. Biol. 2012;78:407–416. doi: 10.1007/s11103-012-9875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods. 2012;9:588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS One. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 66.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat. Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 68.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “Open-Source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2010;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herrmann F, Garriga-Canut M, Baumstark R, Fajardo-Sanchez E, Cotterell J, Minoche A, Himmelbauer H, Isalan M. p53 Gene repair with zinc finger nucleases optimised by yeast 1-hybrid and validated by Solexa sequencing. PLoS One. 2011;6:e20913. doi: 10.1371/journal.pone.0020913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen K, Coussens MJ, Sago J, Subramanian S, Gjoka M, Briner D. Genome editing with CompoZr custom zinc finger nucleases (ZFNs) J. Vis. Exp. 2012:e3304. doi: 10.3791/3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mineta Y, Okamoto T, Takenaka K, Doi N, Aoyama Y, Sera T. Enhanced cleavage of double-stranded DNA by artificial zinc-finger nuclease sandwiched between two zinc-finger proteins. Biochemistry. 2008;47:12257–12259. doi: 10.1021/bi801800k. [DOI] [PubMed] [Google Scholar]
- 75.Mino T, Aoyama Y, Sera T. Efficient double-stranded DNA cleavage by artificial zinc-finger nucleases composed of one zinc-finger protein and a single-chain FokI dimer. J. Biotechnol. 2009;140:156–161. doi: 10.1016/j.jbiotec.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Mori T, Kagatsume I, Shinomiya K, Aoyama Y, Sera T. Sandwiched zinc-finger nucleases harboring a single-chain FokI dimer as a DNA-cleavage domain. Biochem. Biophys. Res. Commun. 2009;390:694–697. doi: 10.1016/j.bbrc.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 77.Fan B, Huang P, Zheng S, Sun Y, Fang C, Sun Z. Assembly and in vitro functional analysis of zinc finger nuclease specific to the 3′ untranslated region of chicken ovalbumin gene. Anim. Biotechnol. 2011;22:211–222. doi: 10.1080/10495398.2011.626885. [DOI] [PubMed] [Google Scholar]
- 78.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc. Natl Acad. Sci. USA. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Isalan M, Klug A, Choo Y. Comprehensive DNA recognition through concerted interactions from adjacent zinc fingers. Biochemistry. 1998;37:12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 81.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 83.Deng D, Yin P, Yan C, Pan X, Gong X, Qi S, Xie T, Mahfouz M, Zhu JK, Yan N, et al. Recognition of methylated DNA by TAL effectors. Cell Res. 2012;22:1502–1504. doi: 10.1038/cr.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, II, Tan W, Penheiter SG, Ma AC, Leung AYH, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]