Abstract
PrePPI (http://bhapp.c2b2.columbia.edu/PrePPI) is a database that combines predicted and experimentally determined protein–protein interactions (PPIs) using a Bayesian framework. Predicted interactions are assigned probabilities of being correct, which are derived from calculated likelihood ratios (LRs) by combining structural, functional, evolutionary and expression information, with the most important contribution coming from structure. Experimentally determined interactions are compiled from a set of public databases that manually collect PPIs from the literature and are also assigned LRs. A final probability is then assigned to every interaction by combining the LRs for both predicted and experimentally determined interactions. The current version of PrePPI contains ∼2 million PPIs that have a probability more than ∼0.1 of which ∼60 000 PPIs for yeast and ∼370 000 PPIs for human are considered high confidence (probability > 0.5). The PrePPI database constitutes an integrated resource that enables users to examine aggregate information on PPIs, including both known and potentially novel interactions, and that provides structural models for many of the PPIs.
INTRODUCTION
Knowledge of protein–protein interactions (PPIs) is essential to understanding cellular regulatory processes. Much effort involving a multitude of methods has been devoted to the determination of direct physical interactions between proteins (1,2). Although most detection methods can only be used for small-scale studies, a few techniques, such as the yeast two-hybrid assays and affinity purification, can be scaled up to determine PPIs in a high-throughput manner (3,4). These high-throughput techniques have been applied to genome-wide studies of PPIs for a number of model organisms, including yeast (5–12), fly (13), worm (14), bacteria (15,16), human (17–19) and, more recently, Arabidopsis (20).
A number of databases have been created to systematically collect and store information on experimentally determined PPIs, including the Munich Information Center for Protein Sequence (MIPS) protein interaction database (21), the database of interacting proteins [DIP, (22)], the protein interaction database [IntAct, (23)], the molecular interaction database [MINT, (24)], the Human Protein Reference Database [HPRD, (25)] and the Biological General Repository for Interaction Datasets [BioGRID, (26)]. To date, hundreds of thousands of PPIs have been stored in these databases that cover hundreds of different organisms and contain interactions determined by tens of different methods (27,28).
Although these databases are crucially valuable resources, they inevitably contain some number of false interactions (false positives) and are largely incomplete in that many interactions are still not annotated (false negatives) (29–31). Although false negatives mainly result from the inherent limitations of different detection methods and incomplete screening of the vast possible interaction space, false positives in these databases can result from errors or ambiguities in experiments (32). In particular, data sets generated from high-throughput methods are estimated to have a much higher error rate than traditional small-scale studies (33). In addition to experimental errors, false-negative and false-positive interactions also result from curation errors. For example, a study of discrepancies between different databases showed that, even for the same set of publications, two databases on average only fully agree on 42% of the interactions and 62% of the proteins (34). The differences were attributed to divergent assignments of organism or splice isoforms, and alternative representations of multiprotein complexes, etc.
Parallel to experimental studies and literature curations, computational predictions have also been used to infer new interactions from indirect clues. Information such as sequence and structural homology, domain–domain interaction profile, genomic context, gene fusion, phylogenetic profile/tree similarity, gene co-expression, function similarity and network topology has been effectively exploited to evaluate the reliabilities of experimentally determined interactions (35,36), and to predict PPIs on a large scale (37–41). Usually, every indirect clue by itself is only a weak PPI predictor, but predictions can be improved by integrating different sources of evidence using a variety of machine learning methods. There have been a number of online databases that store PPIs predicted from these integrative methods, such as STRING (42), Predictome (43), OPHID (44) and its replacement I2D, IntNetDB (45) and PIPs (46). These databases have their own limitations, and it should be noted that, owing to the nature of many prediction methods, many of the predicted interactions are often more indicative of protein functional associations than of direct physical interactions.
Recently, we described a PPI prediction method (PrePPI) that is largely based on 3D protein structural information (47). We showed that, with the exploitation of homology models and remote geometric relationships, structural information can be used to accurately predict PPIs on a genome-wide scale. The further integration of structural with other functional clues yields prediction performance comparable with high-throughput experiments. Experimental tests of a number of predictions demonstrate the ability of the structure-based algorithm to identify novel unsuspected PPIs of significant biological interest.
Given the inconsistent levels of reliability and lack of complete overlap between different PPI databases, a resource that integrates different sources of information and that reports an appropriate measure of reliability should be extremely valuable. In this article, we describe the PrePPI database that contains interactions predicted from our structure-based integrative method, and also includes interactions compiled from a set of public databases that manually curate experimentally determined PPIs from the literature. A probability for each interaction is calculated using a Bayesian framework as described later in the text.
THE PrePPI DATABASE: DATA SOURCES
Predicted interactions
Predicted interactions in the PrePPI database are generated by our structure-based integrative PPI prediction method that combines structural modeling with other genomic, evolutionary and functional clues (47). Briefly, for a pair of proteins of interest, we first search for representative structures of the query proteins in the PDB and homology model databases, and then use these to search for structural neighbors of each protein. A protein–protein complex found in the Protein Quaternary Structure database or Protein Data Bank is used as a ‘template’ for the interaction whenever it contains a pair of interacting chains that are structural neighbors of the respective query proteins. We then construct a model by superposing the individual subunits on their corresponding structural neighbors in the template complex and calculate a likelihood ratio (LR) for each model to represent a true interaction using a Bayesian network trained on a positive and a negative interaction reference set. We finally combine the structure-derived LR with non-structural evidence associated with the query proteins using a naïve Bayesian classifier.
Our analyses show that the performance of the prediction method is comparable with high-throughput studies, and that this is primarily due to the large-scale use of structural information made possible by the use of homology models and looking broadly across protein structure space for structure/function relationships. To put this in perspective, using structure alone we build structural models for ∼2.4 million and 36 million yeast and human interactions, respectively.
Experimentally determined interactions
We collected PPIs from six publicly available databases (MIPS, DIP, IntAct, MINT, HPRD and BioGRID) and obtained 117 803 interactions for yeast and 82 060 interactions for human. We mapped protein identifiers from different databases to UniProt accession numbers and used pairs of accession numbers as the unique identifiers of all PPIs. Different databases contain different numbers of false-positive and false-negative interactions that are due to both experimental and curation errors. We have used Bayesian statistics to calculate an LR for database interactions as follows. We used a positive reference set that contains 11 851 yeast interactions and 7409 human interactions that have more than one supporting publication, and a negative reference set constructed by pairing proteins located in different cellular compartments (47). We assigned each of these interactions to one of seven categories and calculated an LR for each category. The first category contains interactions that are present in multiple databases, and the other six contain interactions present in exclusively one of the databases listed earlier in the text. In this way, we obtain an objective evaluation that accounts for both experimental and curation quality.
Combining the LRs for predicted and experimentally determined interactions
An advantage of using a Bayesian framework to calculate an LR for each database is that we can easily combine experimentally determined interactions with computationally predicted interactions. Because the two are weakly correlated, we use a naïve Bayesian classifier to combine them by simply multiplying the two LR scores to obtain a combined LR score for each interaction.
In the PrePPI database, we have scaled the combined LR to a probability using the following equation:
| (1) |
We use an LRcutoff of 600, which roughly corresponds to a false-positive rate of 0.001, based on the assumption that the probability that an interaction of LR 600 is true is 0.5 (47,48).
The PrePPI database now contains ∼2 million PPIs with a probability >0.1. Of these, 61 720 PPIs for yeast and 372 545 PPIs for human have a probability >0.5.
THE PrePPI DATABASE: WEB INTERFACE
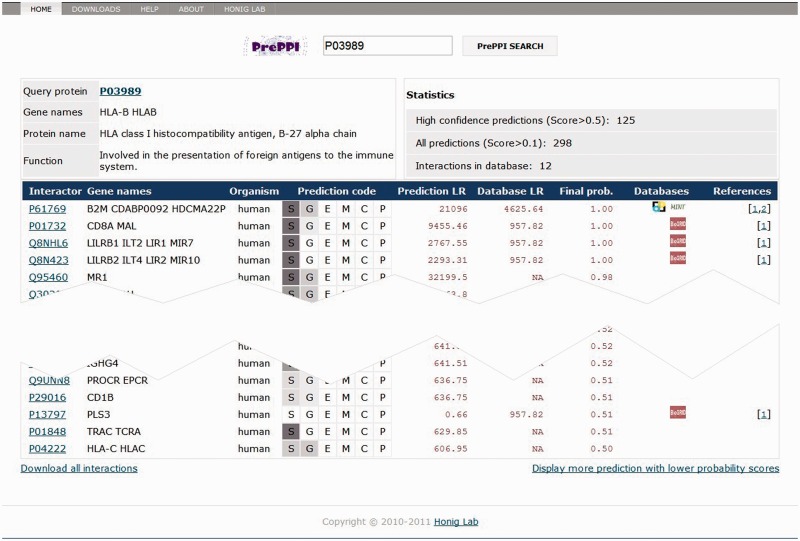
The PrePPI database can be queried through the UniProt accession number (e.g. P03989), gene name (e.g. PRNP) or protein name (e.g. Histone H2A) of a gene or protein. The server will return a description of the query protein, the number of proteins it interacts with and a table with detailed information about each interaction (Figure 1). Each row of the table lists proteins predicted to interact with the query, the sources of information used in the prediction, different LRs and the final combined probability, as well as whether the interaction has been documented in databases or in the literature.
Figure 1.
The PrePPI page of predicted protein–protein interactions for query protein P03989.
The sources of information used in the prediction are represented by their ‘prediction codes’. Details on different types of information can be found in the ‘Help’ page of the web server. The ‘Prediction LR’ column shows the LR obtained from the Bayesian network that combines the different sources of structural and non-structural evidence for the interaction represented by their prediction codes [see (47) for details on the types of evidence used]. We also calculate a ‘database LR’ as described earlier and combine this with the prediction LR to get a final LR, which is shown in the table as a probability (Final prob.) determined from Equation 1. If an interaction has been previously documented, we put the corresponding database symbols in the seventh column and the PubMed links to the description of the relevant experiments in the eighth column.
Interactions are ordered according to their final probabilities. By default, we only show the high confidence predictions (final probability >0.5), but predictions with lower probabilities can be viewed by clicking the link at the bottom right. All interactions for the query protein can be downloaded by clicking the link at the bottom left.
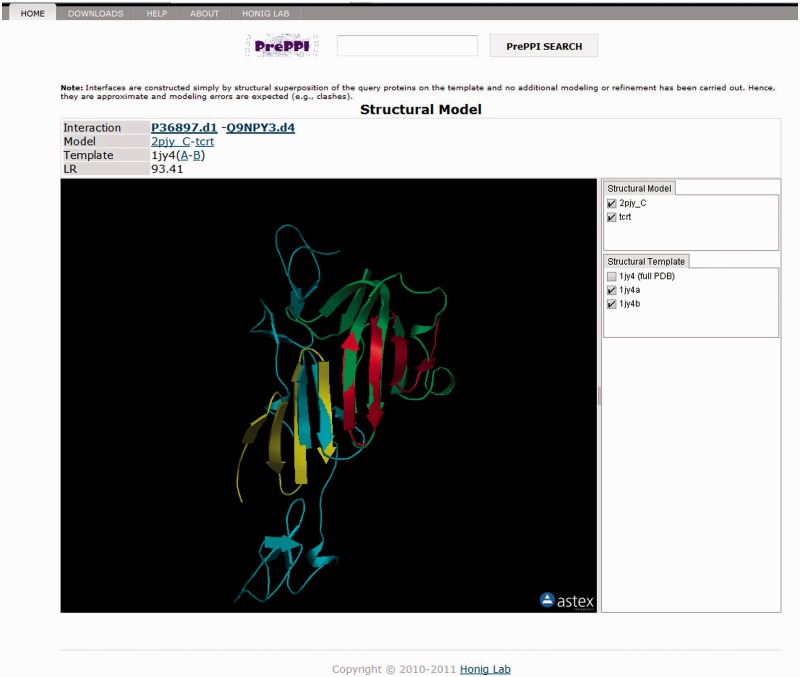
A unique feature of the PrePPI database is the availability of structural interaction models for those PPIs predicted from our structural modeling algorithm. Figure 2 shows an example of an interaction model built for the human TGF-β receptor type-1 (P36897) and the complement component C1q receptor (Q9NPY3), using a homology model from Skybase (49) for Q9NPY3 and exploiting the remote structural relationship between these monomer structures and a designed protein that forms a homodimer (50). Users can investigate the interaction model and generate experimentally testable hypotheses for how the two proteins interact. It is important to emphasize that no structural refinement of PrePPI models is carried out, so they may contain physically unrealistic features such as steric clashes. The structure-based LR for the model is shown in the viewer and, together with the reasonableness of the model itself, should be considered when evaluating its biological relevance and when deciding whether some form of structural refinement might be of value.
Figure 2.
The structural interaction model for TGF-β receptor type I (green, UniProt ID P36897) and complement component C1q receptor (cyan, UniProt ID Q9NPY3) based on the structure of a designed protein (gold and red for A and B chains, respectively, of PDB file 1jy4).
CAVEATS AND PLANS
The goal of PrePPI is to generate testable hypotheses derived in part from structure, but its use should be seen, in our opinion, as an early step in the process of biological discovery. PrePPI is under constant development, but at this stage, it is worth pointing out a number of caveats. First, although we have shown that the structure-based LR can account for specificity in the sense that it can differentiate closely related structural domains that form complexes from those that do not [see Figure S15 in the supplemental material of (47)], the methods used are not perfect and predictions should be considered carefully in the context of any additional data that might be available (for example, the highest scoring predictions may be paralogs that appear in different cellular compartments). As discussed earlier in the text, other problems may arise from the fact that we do not attempt to evaluate the 3D model of a putative complex beyond scoring of the interface (47) so that in many cases the model may appear physically unrealistic. Ideally, it will be possible to address such issues automatically through, for example, the use of orthology databases or refinement of side chains, loops and relative domain orientations. We plan to implement such features in future versions of PrePPI. However, because PrePPI evaluates billions of interaction models (47), structural refinement would have to be carried out in a later filtering step, perhaps motivated by biological interest. At this stage, we have chosen to present all high probability predictions with the expectation that a thoughtful user will be able to recognize obvious false positives using the information available on the server itself, in external databases or in the biological literature.
Finally we note that a high probability PrePPI prediction for an interaction says nothing about the oligomerization state of the proteins involved. Our goal at this stage is to assign a probability for an interaction between two proteins to occur and provide an initial model of where an interface might be located. Again, our hope is that the interested user will be able to use the information provided in the PrePPI database as a basis for new experimental and computational efforts on a particular system of interest.
CONCLUSION
The PrePPI database differs from other PPI databases based on the following four novel features: (i) PrePPI provides structural information for many more interactions than has previously been possible using structure-enabled approaches and databases (51–53); (ii) the predicted PPIs in PrePPI are obtained by combining structural and non-structural information; (iii) the PrePPI database contains integrative information of PPIs from major PPI databases and provides a Bayesian measure as to the confidence level of these interactions; and (iv) the PrePPI database assigns a single probability for each interaction using a Bayesian framework that combines quantitative results based on computational predictions with evidence contained in publicly available databases. PrePPI now offers a comprehensive source of PPI information for the yeast and human genomes and will soon be expanded to other model organisms.
FUNDING
National Institutes of Health [GM030518, GM094597, CA121852]. Funding of the open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol. Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput. Biol. 2007;3:e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parrish JR, Gulyas KD, Finley RL., Jr Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 2006;17:387–393. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Vasilescu J, Figeys D. Mapping protein-protein interactions by mass spectrometry. Curr. Opin. Biotechnol. 2006;17:394–399. doi: 10.1016/j.copbio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 6.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 8.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 9.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 10.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 13.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 16.Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 17.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 18.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mewes HW, Albermann K, Heumann K, Liebl S, Pfeiffer F. MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 1997;25:28–30. doi: 10.1093/nar/25.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the molecular interaction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehne B, Schlitt T. Protein-protein interaction databases: keeping up with growing interactomes. Hum. Genomics. 2009;3:291–297. doi: 10.1186/1479-7364-3-3-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai J, Rohl C, Price Y, Fischer TB, Paczkowsk M, Zette MF. Cataloging the relationships between proteins. Mol. Biotechnol. 2006;34:69–93. doi: 10.1385/MB:34:1:69. [DOI] [PubMed] [Google Scholar]
- 29.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 30.Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Sprinzak E, Sattath S, Margalit H. How reliable are experimental protein-protein interaction data? J. Mol. Biol. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 33.Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turinsky AL, Razick S, Turner B, Donaldson IM, Wodak SJ. Literature curation of protein interactions: measuring agreement across major public databases. Database. 2010;2010:baq026. doi: 10.1093/database/baq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 36.Bader JS, Chaudhuri A, Rothberg JM, Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 37.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part II. Computational methods to predict protein and domain interaction partners. PLoS Comput. Biol. 2007;3:e43. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valencia A, Pazos F. Computational methods for the prediction of protein interactions. Curr. Opin. Struct. Biol. 2002;12:368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 39.Salwinski L, Eisenberg D. Computational methods of analysis of protein-protein interactions. Curr. Opin. Struct. Biol. 2003;13:377–382. doi: 10.1016/s0959-440x(03)00070-8. [DOI] [PubMed] [Google Scholar]
- 40.Szilagyi A, Grimm V, Arakaki AK, Skolnick J. Prediction of physical protein-protein interactions. Phys. Biol. 2005;2:S1–S16. doi: 10.1088/1478-3975/2/2/S01. [DOI] [PubMed] [Google Scholar]
- 41.Musso GA, Zhang Z, Emili A. Experimental and computational procedures for the assessment of protein complexes on a genome-wide scale. Chem. Rev. 2007;107:3585–3600. doi: 10.1021/cr0682857. [DOI] [PubMed] [Google Scholar]
- 42.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellor JC, Yanai I, Clodfelter KH, Mintseris J, DeLisi C. Predictome: a database of putative functional links between proteins. Nucleic Acids Res. 2002;30:306–309. doi: 10.1093/nar/30.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 45.Xia K, Dong D, Han J-D. IntNetDB v1.0: an integrated protein-protein interaction network database generated by a probabilistic model. BMC Bioinformatics. 2006;7:508. doi: 10.1186/1471-2105-7-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDowall MD, Scott MS, Barton GJ. PIPs: human protein–protein interaction prediction database. Nucleic Acids Res. 2009;37:D651–D656. doi: 10.1093/nar/gkn870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 49.Mirkovic N, Li Z, Parnassa A, Murray D. Strategies for high-throughput comparative modeling: applications to leverage analysis in structural genomics and protein family organization. Proteins. 2007;66:766–777. doi: 10.1002/prot.21191. [DOI] [PubMed] [Google Scholar]
- 50.Venkatraman J, Nagana Gowda GA, Balaram P. Design and construction of an open multistranded β-sheet polypeptide stabilized by a disulfide bridge. J. Am. Chem. Soc. 2002;124:4987–4994. doi: 10.1021/ja0174276. [DOI] [PubMed] [Google Scholar]
- 51.Stein A, Céol A, Aloy P. 3did: identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2011;39:D718–D723. doi: 10.1093/nar/gkq962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lo Y-S, Chen Y-C, Yang J-M. 3D-interologs: an evolution database of physical protein- protein interactions across multiple genomes. BMC Genomics. 2010;11:S7. doi: 10.1186/1471-2164-11-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]