Abstract
The Epigenomics resource at the National Center for Biotechnology Information (NCBI) has been created to serve as a comprehensive public repository for whole-genome epigenetic data sets (www.ncbi.nlm.nih.gov/epigenomics). We have constructed this resource by selecting the subset of epigenetics-specific data from the Gene Expression Omnibus (GEO) database and then subjecting them to further review and annotation. Associated data tracks can be viewed using popular genome browsers or downloaded for local analysis. We have performed extensive user testing throughout the development of this resource, and new features and improvements are continuously being implemented based on the results. We have made substantial usability improvements to user interfaces, enhanced functionality, made identification of data tracks of interest easier and created new tools for preliminary data analyses. Additionally, we have made efforts to enhance the integration between the Epigenomics resource and other NCBI databases, including the Gene database and PubMed. Data holdings have also increased dramatically since the initial publication describing the NCBI Epigenomics resource and currently consist of >3700 viewable and downloadable data tracks from 955 biological sources encompassing five well-studied species. This updated manuscript highlights these changes and improvements.
INTRODUCTION
The field of epigenetics is garnering increasing amounts of interest in the scientific community. Epigenetics refers to the study of stable, often heritable, changes that influence gene expression that are not mediated by DNA sequence (1,2). Epigenetic mechanisms play crucial roles in chromatin state regulation, thereby influencing processes such as gene expression, DNA repair, and recombination. Although individual epigenetic features are tied to specific genomic locations and can be stably inherited through many rounds of cell division, these epigenetic features can be modified, or erased in response to developmental cues or external and environmental stimuli (3–6). Just as these epigenetic mechanisms strongly influence development and cellular processes, defects in these mechanisms can prove to be quite deleterious. It is now known that certain defects in epigenetic regulation can be linked to instances of human disease, including developmental defects, metabolic disorders and cancer (7–9). Additionally, links are being uncovered between the epigenome and more common complex diseases including psychosis, diabetes and asthma. A better understanding of the epigenomic factors that contribute to these disease processes will lead to additional strategies for treatment in the future. It has become a major driving force behind epigenomics research (10–13).
Epigenetic modifications are varied and diverse yet fall into four major classes: post-translational modification of histone proteins, chromatin conformation/accessibility, DNA modification and non-coding regulatory RNA (3,14). These mechanisms have been intensely studied and are well characterized. Covalent modification of histone proteins can induce or relax the packaging constraints of chromatin (15). Modification of DNA, specifically methylation of cytosine, is crucial for processes such as DNA imprinting, X-chromosome inactivation and long range silencing of genomic regions (3,16). Other modified forms of cytosine have more recently been discovered that show distinct genomic localizations and functions. These include 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine (17). Non-coding RNA molecules can interact with specific target mRNAs and trigger a cascade of events resulting in specific mRNA degradation (18–20). Chromatin accessibility and nucleosome positioning have also been determined to serve as epigenetic mechanisms. It is not uncommon to find elements that regulate gene expression (e.g. promoters, enhancers, insulators) in regions of the genome that are maintained as ‘open’ or accessible. These accessible regions can serve as binding sites for chromatin-modifying enzymes and other protein factors (21). These epigenetic mechanisms often act in concert for more complex levels of regulation. For example, it has been observed that small non-coding RNA molecules can participate in directing DNA methylation, and that enhancer elements, often found in regions where chromatin is maintained as accessible, can encode for small non-coding RNA molecules themselves (18–20,22).
The distribution of these epigenetic features throughout the genome constitutes what can be considered the cellular ‘epigenome’. The epigenome, unlike the genome itself, is dynamic, and localization of these epigenetic features is influenced by cell or tissue type, age, exposure to environmental stimuli or countless other factors. These factors make defining an organism’s singular epigenome a daunting, if not impossible, task. Yet, owing to the complex and important roles that epigenetic phenomena play in human health and development, efforts to understand the human epigenome are underway. To address this, the NIH launched the Roadmap Epigenomics Project in 2007 (23). One of the goals of this project is to combine whole genome epigenetic analysis with high throughput sequencing to create a series of publically available reference epigenome maps. These maps will encompass a wide array of cell lines, cell and tissue types from individuals at various developmental stages and health states. Other large scale efforts to map epigenetic features and gene regulatory elements are currently ongoing. This includes the ENCODE (ENCyclopedia of DNA Elements) project, Mouse ENCODE and modENCODE project for model organisms (24–27). The National Center for Biotechnology Information (NCBI) Epigenomics database was created as a repository for these data as well as from other independent labs not involved in these project initiatives (28). In this article, we will describe new features, improvements and current holdings in this growing resource.
THE EPIGENOMICS DATABASE
The organizational framework for the database involves studies, samples, experiments and genome tracks. At the lowest level, a genome track is a representation of a signal or annotation in the coordinate systems of a specific genome assembly. At present, all of the tracks in the database are molecular abundance graphs (e.g., enrichment of a modified histone). However, the database is designed to capture additional types of tracks (e.g., peak calls or chromatin state maps) once they become more widely available. An experiment refers to the laboratory assay that generated the raw data that were used to construct a track. It is possible for there to be multiple tracks for one experiment, either because several types of tracks were made or because there are variants for different assemblies of the genome. A sample refers to the biological material that was used for the experiment, which is an essential unit for allowing all of the results from one isolate to be grouped together. Finally, a study is a group of experiments with a common set of scientific aims.
In building the Epigenomics resource, data are selected from Gene Expression Omnibus (GEO) database and subjected to additional processing and tracking. GEO is a database of large-scale molecular abundance data generated for functional genomics studies (29). Because most of the experiments in scope for epigenomics are based on sequencing methodologies, there are often companion submissions to the Sequence Read Archive (SRA) database (30). Genome tracks may be attached to GEO submissions as supplementary files in a wide variety of formats. For molecular abundance graphs, we currently accept WIG and bigWig files. These data are subjected to computational analysis to identify the likely genome assembly and to ensure that all genome coordinates are in a valid range. Tracks are given accession numbers, and changes are tracked using revision numbers and update dates. In some cases, submitted tracks that had been constructed using an older genome assembly will be remapped to reflect the current state of the genome. In this case, the derived track has a separate accession number and revision chain to keep it distinct from the original submission. Incidentally, for records in Epigenomics, links are provided to original submitted records and data at GEO and SRA. These raw unprocessed data can be accessed and downloaded by users who are interested in performing their own analyses.
Layered on top of the basic track data are metadata in the form of controlled key terms and relationships with records in other databases. Many of these attributes pertain to properties of the biological material, such as cell type, differentiation state, health status and so forth. Others are properties of the experiment, such as assay type and (where relevant) specific antibodies. Relationships between experiment records and the original data in GEO and SRA are captured. Some of the larger studies may have links to a new NCBI database called BioProject, which describes the project aims and provides links to associated data (31). Finally, nearly all studies will ultimately have one or more publications, which are captured as links to PubMed citations and (where applicable) full-text articles in PubMed Central (PMC).
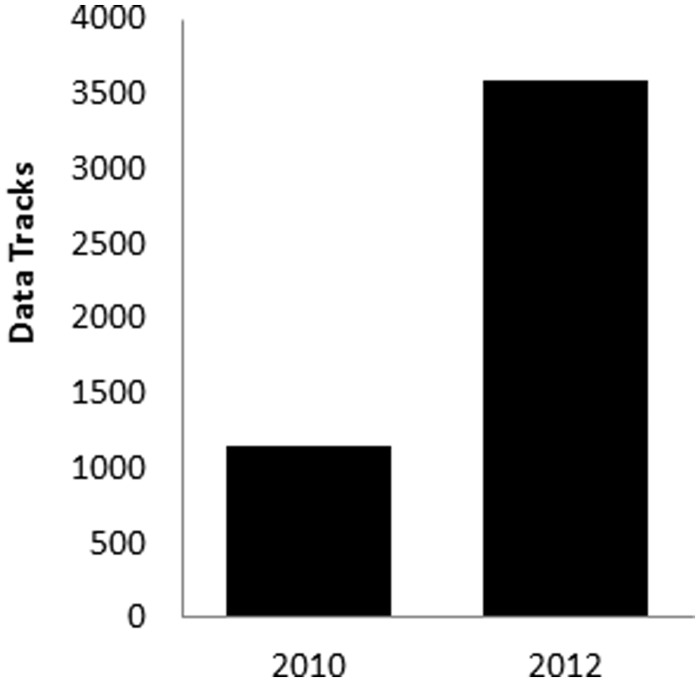
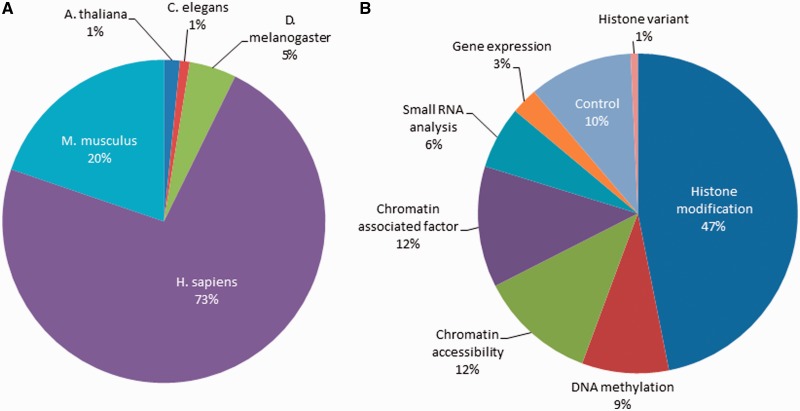
The database holdings have grown dramatically over the past few years. As shown in Figure 1, the total number of database records has increased >3-fold since our previous report 2 years ago (values are from September of the indicated year). As of this writing, the database contains 3708 genome tracks sourced from 955 biological samples. These data come from five well-studied species (Figure 2A). Given the large output from the Roadmap Epigenomics and ENCODE projects, it is not surprising that the bulk of the records (73%) are of human origin. There is also a significant amount of data from mouse (20%), but smaller amounts from the other model organisms. Data tracks reflect a variety of assay types (Figure 2B), dominated by histone modifications (47% of the total), but also including DNA methylation, chromatin accessibility and various chromatin-associated factors (including RNA polymerase, transcription factors and various histone-modifying enzymes). Although not epigenetic per se, gene expression—either at the level of mRNA or small non-coding RNAs—is often assayed along with epigenetic marks in order to advance understanding of gene regulatory networks. Finally, various sorts of controls are often included in submissions (input control, antibody control), and they are included in the resource to allow for use in normalization or quality assessment.
Figure 1.
Track data in the NCBI Epigenomics resource over time. Holdings have increased by >3-fold in the time period spanning from 2010 to 2012. Currently there are 3708 tracks available.
Figure 2.
Composition of track holdings in the NCBI Epigenomics resource. (A) Percentage of holdings by species. Species include Homo sapiens (H. sapiens), Mus musculus (M. musculus), Arabidopsis thaliana (A. thaliana), Drosophila melanogaster (D. melanogaster) and Caenorhabditis elegans (C. elegans). (B) Percentage of holdings by assay type. Assay types include histone modifications, DNA methylation, chromatin accessibility, various chromatin-associated factors (including RNA polymerase, transcription factors and various histone-modifying enzymes) and small RNA and gene expression.
THE EPIGENOMICS WEBSITE
All content of the database is made publically available through the Epigenomics resource on the NCBI website (www.ncbi.nlm.nih.gov/epigenomics/). In the course of the past 2 years, we have made incremental improvements to the resource-based feedback from the user community, together with our own usability testing and web log analysis. In addition, the site has benefited from general enhancements of the NCBI search interface, including faceted searching, spelling correction and synonymy mapping (e.g. mapping ‘ESC’ to ‘embryonic stem cell’).
An example of an improvement that came from user feedback is the extension of the Sample Browser interface to work with experiment records. This tool lists database records in a tabular (spreadsheet-like) format, with columns corresponding to various biological and experimental attributes. Now, users can easily switch back-and-forth between browsing these two classes of documents while retaining the familiar features—filtering the table content, sorting on any of the columns and exporting the information in a spreadsheet compatible format. Additionally, user feedback has shaped the specifics of the filtering options and choices of which columns to show by default (although users are free to change these defaults). The Sample Browser also serves as a starting point for other tools and features of the site, such as saving records of interest in user-defined collections, bulk data downloading and graphical visualization using the NCBI genome viewer [with a link to the corresponding view on the University of California, Santa Cruz (UCSC) Genome Browser].
Another line of enhancements was driven by web analytics, starting with the observation that a large fraction of the free-text searches performed on the site did not yield any results. From inspection of a few hundred such queries, we concluded that about half of them were gene symbols or other words aimed at finding specific genes or gene families. Because all of the records in the database represent some form of whole-genome analysis, they are not indexed by gene symbol. To address this problem, we developed a search enhancement that would check for gene symbols in the query text and—when found—generate a ‘featured result’ with a direct link to a graphical genome view centered on the gene of interest. In these views, a selected set of reference tracks is displayed, leaving it to the user to substitute other tracks of interest. Follow-up analysis of the logs showed that ∼22% of all site visitors made use of the feature at some time during their session and ∼30% of all empty results were ‘rescued’ by directing the user to some useful content. Clearly, genes are useful starting points for researchers, so we worked with the staff of the NCBI Gene database to add links to the Epigenomics genome views on records for human genes. When the updated views were deployed, there was a concomitant doubling of overall usage of the Epigenomics resource and a 4-fold increase in the number of genome views. To make the genome views more useful, we have added annotation tracks for CpG islands and clinically relevant sequence variations.
We have developed a comparison tool that is aimed at identifying genes that have undergone some sort of change between one biological state and another—say, before and after differentiation. The tool works by identifying clustered genomic positions that show the most difference in one track compared with another and then connecting them to genes based on proximity. A short list of the genes with the most differences is generated, and the result includes small thumbnail graphics depicting the signal at that locus for each of the two samples. To give the user a general flavour for the kinds of genes involved, functional terms that occur frequently in the top genes are listed [terms come from the Genome Ontology (GO) or names of pathways in the NCBI BioSystems database].
A recently developed feature provides users with the ability to upload custom data tracks. To maintain privacy and to support long-term data storage, it is necessary for users to have an account and be signed in before uploading tracks. Accounts are free and may be established in the My NCBI part of the site (be aware that quotas on number of uploaded tracks per account may be adjusted over time depending on demand). This upload functionality supports popular file formats including BED and WIG. Data can be uploaded from a local file or a public URL. Along with the data track itself, a small number of metadata attributes may be entered if desired. On completion of the uploading process, custom tracks are listed in the Experiment Browser interface and may be viewed graphically alongside public tracks.
CONCLUSION
The Epigenomics database at NCBI was established to serve as a public resource for epigenomic data sets. Data have been collected from both large scale studies such as the NIH Roadmap Epigenomics project, ENCODE and modENCODE and from smaller single laboratory studies. The holdings in Epigenomics have increased more than 3-fold over the previous 2 years, and continue to grow. We have implemented new tools and new features in response to user feedback including the ability to browse the database at the experimental level, compare and upload data tracks. We anticipate a continuing growth of data holdings, development of new tools and improvement of general usability in the future. It is our goal to continue providing a comprehensive public resource for epigenomic datasets that gives users, with varying degrees of knowledge in the field, the ability to analyse and explore epigenomic data sets.
FUNDING
Funding for open access charge: The Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge and thank Tanya Barrett and Alexandra Soboleva for their input into establishing the Epigenomics resource. The authors would also like to thank members of other NCBI teams managing GEO, Sequence Viewer and Log Analysis for essential contributions to the Epigenomics resource.
REFERENCES
- 1.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 2.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 5.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 6.Roloff TC, Nuber UA. Chromatin, epigenetics and stem cells. Eur. J. Cell Biol. 2005;84:123–135. doi: 10.1016/j.ejcb.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem. Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 10.Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu. Rev. Pharmacol. Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- 11.Ptak C, Petronis A. Epigenetic approaches to psychiatric disorders. Dialogues Clin. Neurosci. 2010;12:25–35. doi: 10.31887/DCNS.2010.12.1/cptak. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durham A, Chou PC, Kirkham P, Adcock IM. Epigenetics in asthma and other inflammatory lung diseases. Epigenomics. 2010;2:523–537. doi: 10.2217/epi.10.27. [DOI] [PubMed] [Google Scholar]
- 13.Villeneuve LM, Reddy MA, Natarajan R. Epigenetics: deciphering its role in diabetes and its chronic complications. Clin. Exp. Pharmacol. Physiol. 2011;38:451–459. doi: 10.1111/j.1440-1681.2011.05497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003;2003:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 15.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 20.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 22.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science, 306, 636–640. [DOI] [PubMed]
- 25.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fingerman IM, McDaniel L, Zhang X, Ratzat W, Hassan T, Jiang Z, Cohen RF, Schuler GD. NCBI Epigenomics: a new public resource for exploring epigenomic data sets. Nucleic Acids Res. 2011;39:D908–D912. doi: 10.1093/nar/gkq1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shumway M, Cochrane G, Sugawara H. Archiving next generation sequencing data. Nucleic Acids Res. 2010;38:D870–D871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]