Abstract
Knowledge of the various interactions between molecules in the cell is crucial for understanding cellular processes in health and disease. Currently available interaction databases, being largely complementary to each other, must be integrated to obtain a comprehensive global map of the different types of interactions. We have previously reported the development of an integrative interaction database called ConsensusPathDB (http://ConsensusPathDB.org) that aims to fulfill this task. In this update article, we report its significant progress in terms of interaction content and web interface tools. ConsensusPathDB has grown mainly due to the integration of 12 further databases; it now contains 215 541 unique interactions and 4601 pathways from overall 30 databases. Binary protein interactions are scored with our confidence assessment tool, IntScore. The ConsensusPathDB web interface allows users to take advantage of these integrated interaction and pathway data in different contexts. Recent developments include pathway analysis of metabolite lists, visualization of functional gene/metabolite sets as overlap graphs, gene set analysis based on protein complexes and induced network modules analysis that connects a list of genes through various interaction types. To facilitate the interactive, visual interpretation of interaction and pathway data, we have re-implemented the graph visualization feature of ConsensusPathDB using the Cytoscape.js library.
INTRODUCTION
A major goal of systems biology is to assemble an exhaustive global map of the functional relationships, or interactions, between physical entities in the cell (genes, proteins, metabolites, etc.) (1). Many methods have been developed to measure such interactions and have been applied to model organisms and to human. Thus, hundreds of thousands of interactions have already been detected, reported in the literature and assembled in interaction databases (2); however, these databases are often complementary and tend to focus on one or a few types of interactions while in reality all the different interaction types coexist in the cell. In order to obtain a global interaction map that reflects biology as completely as possible, subject to the currently available interaction knowledge, many available interaction resources have to be considered. The heterogeneity of databases in terms of interaction type, data model and data exchange format complicates their integration. To facilitate the exchange and integration of data from different resources, standard file formats such as PSI-MI (3) and BioPAX (4), and respective platforms for data exchange such as PSICQUIC (5) and Pathway Commons (6) have been developed. However, not all interaction resources have adopted standard formats, e.g. because they are not compatible with the data model of the respective resource. Despite these hurdles, we have developed a database called ConsensusPathDB that integrates different types of interactions from numerous resources into a seamless global network (7,8). In this network, physical entities (genes, proteins, metabolites, etc.) from different sources are matched depending on their accession numbers and interactions are matched depending on their participants to reduce data redundancy. The web interface of ConsensusPathDB aims to serve as a one-stop shop for searching, visualizing and retrieving the integrated interaction data, as well as for tools that use these data for interaction- and pathway-centric analysis of genes, proteins and metabolites (resulting, e.g. from large-scale transcriptomics, proteomics or metabolomics experiments). In this database update article, we report the most significant recent advancements of ConsensusPathDB in terms of human interaction content and web interface functionalities. In addition to human data, ConsensusPathDB instances exist for interaction and pathway data from the model organisms, mouse and yeast.
DATABASE CONTENT UPDATE
Since our last report on ConsensusPathDB (8), the database has grown both in terms of different types of interactions supported and in terms of source databases (that is databases whose interaction data are integrated in ConsensusPathDB). Newly integrated interaction types comprise genetic interactions and drug–target interactions in addition to the already supported types (protein–protein interactions, biochemical reactions—metabolic and signaling—as well as gene regulatory interactions). Although human genetic interaction data are currently scarce and there are only 265 such interactions in the latest ConsensusPadthDB version [originating from BioGRID (9)], their number is expected to increase in the future. On the other hand, there are bulks of drug–target interaction data extracted from the literature into several dedicated databases. There are currently 33 081 drug–target interactions in ConsensusPathDB that originate from four such databases.
The number of source databases integrated in ConsensusPathDB has grown over the last 2 years since our last report (8) from 18 to 30 databases. The newly integrated resources are BIND (protein–protein interactions) (10), DrugBank (drug–target interactions) (11), InnateDB (protein–protein, biochemical and gene regulatory interactions) (12), MatrixDB (protein–protein interactions) (13), PDZBase (protein–protein interactions) (14), PhosphoPOINT (protein–protein and biochemical interactions) (15), PhosphoSitePlus (biochemical interactions) (16), PINdb (protein–protein interactions) (17), SignaLink (biochemical pathways) (18), SMPDB (biochemical pathways) (19), TTD (drug–target interactions) (20) and WikiPathways (biochemical pathways) (21). Drug–target interactions have been additionally extracted from the previously integrated databases KEGG (22) and PharmGKB (23). Although we do not curate primary datasets, we have integrated a recently published, large-scale spliceosomal protein–protein interaction network obtained through yeast two-hybrid screening from our own research (24).
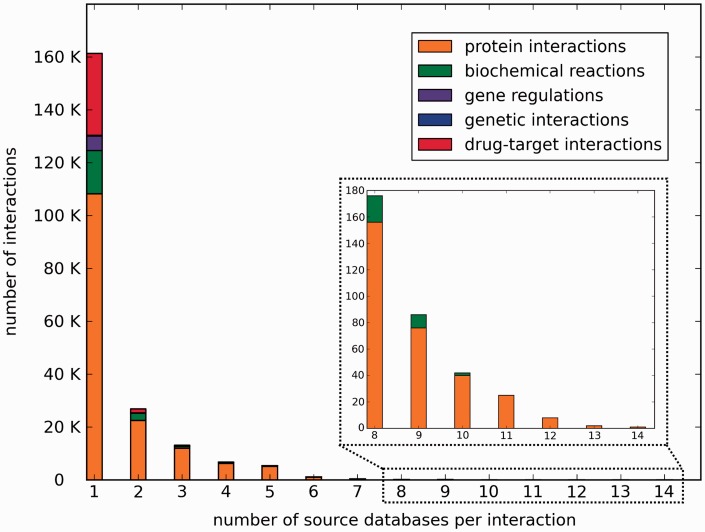
The number of unique interactions stored in ConsensusPathDB has grown in the last 2 years from 155 432 to 215 541 interactions, in part because of the integration of new databases and in part because the content of the previously included resources has grown. Analysis of the total number of source databases per interaction in ConsensusPathDB shows that the respective distribution is right-skewed, with most of the interactions (161 396 interactions, 75%) originating from a single-source database (Figure 1). These results evidence that currently available databases are highly complementary [in agreement with other reports in the literature, e.g. refs. (25) and (26)] and, importantly, that the integrated interaction map present in ConsensusPathDB has not saturated yet. This underlines the importance of further interaction data integration. To rule out effects from missed interaction mappings due to technical issues (e.g. missing accession number annotation of interaction participants), we repeated the analysis considering only those interactions with unambiguously identifiable participants. This analysis showed very similar trends (data not shown).
Figure 1.
Histogram of the number of source databases per interaction in ConsensusPathDB.
Apart from extending the quantity of the ConsensusPathDB content, we have also taken measures for assessing its quality. Interactions stored in public resources are known to be of different confidence. Reportedly, considerable fractions of the available protein–protein interaction data may result from experimental or literature mining errors (26,27). Thus, we have assessed the confidence of binary protein–protein interactions stored in ConsensusPathDB. This was done using an integrative approach that exploits network-topological features and annotation features to derive confidence scores for each individual interaction. Among the network-topological methods integrated in the approach is a parameter-free, reference data-independent method for scoring large binary interaction networks called CAPPIC, which was developed by us (28). The integrative approach has been implemented as a web tool called IntScore (http://intscore.molgen.mpg.de) (29), which was applied to the ConsensusPathDB protein–protein interaction network (Supplementary Methods). Notably, the protein–protein interactions in ConsensusPathDB are only scored and not filtered. The available scores are displayed in the web interface and can be used as a filtering criterion by the users.
WEB INTERFACE FEATURES UPDATE
Pathway analysis of metabolite lists
Over the past decade, pathway over-representation/enrichment analysis of gene lists has proven a very useful tool for interpreting large-scale transcriptomics and proteomics data (30). Such analysis is able to pinpoint biochemical pathways that are dysregulated and may have a causative relationship to the phenotype under study or act as conductors of biological signal leading to it. With the possibility to measure the cellular concentrations of a panel of metabolites provided by state-of-the-art technologies, metabolite signatures for more and more phenotypes are being generated (31). Like abnormal gene expression, abnormal metabolite concentrations can also provide clues about potentially dysregulated metabolic or signaling pathways in the samples under study. To facilitate the analysis of metabolomics data on the pathway level, the web interface of ConsensusPathDB now provides pathway over-representation and enrichment analysis functionality for user-specified metabolite lists. It exploits the fact that most of the pathways stored in our database (3321 out of 4601 pathways, 72%) contain metabolites additionally to genes. Statistical tests are performed with the user-specified metabolite input that are analogous to those described previously in the context of gene set analysis to identify candidate phenotype-associated pathways (7). Although several tools for pathway-based evaluation of metabolite lists are already available (32–34), ConsensusPathDB has the advantage of possessing a rich pathway repertoire collected from 12 resources for biochemical pathways. Moreover, if the user has both transcriptomics/proteomics and metabolomics data from a particular phenotype at hand, ConsensusPathDB can serve as a one-stop shop for analyzing these data based on the same set of pathways. This will save the user time and effort needed to get familiar with two separate tools for the analysis of the different data types, which will besides be typically based on different sets of pathways.
Visualization of functional gene/metabolite sets as overlap graphs
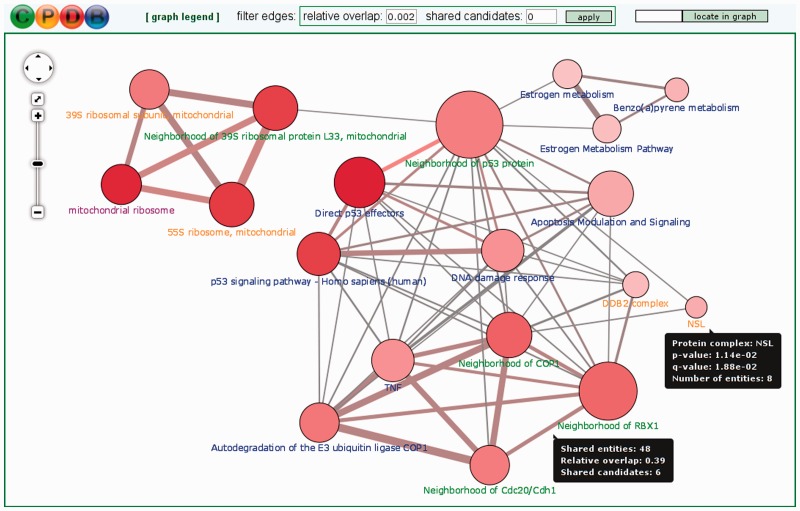
The typical output of most tools for gene/metabolite set over-representation/enrichment analysis is a table where predefined functional gene/metabolite sets (e.g. pathways) are listed, ranked according to some statistical measure of association with the user-specified input (most often a P-value). However, the functional sets often overlap with each other to some extent—for example, they may stand in a hierarchical relationship to each other [like Reactome pathways (35) or Gene Ontology categories (36)] or may share key elements. Thus, to facilitate the visual interpretation of over-representation/enrichment analysis results, we have introduced in ConsensusPathDB the possibility to visualize the resulting functional gene/metabolite sets (pathways, neighborhood-based entity sets (NESTs) (8), Gene Ontology categories and protein complexes) as overlap graphs (Figure 2). In these graphs, each node represents a separate predefined functional set whose member list size (i.e. number of genes/metabolites contained) and P-values are encoded as node size and color, respectively. Two nodes are connected by an edge if the according functional sets share members (genes/metabolites). The edge width reflects the relative overlap calculated with the Fowlkes–Mallows index (37) from the number of shared members and the sizes of the two gene/metabolite sets. The edge color encodes the number of shared members that are also found in the user’s input (denoted ‘shared candidates’). The user can click on the nodes and edges of the overlap graph to view a list of the pertinent genes/metabolites. The visual representation helps the user to quickly identify related biological processes that together show a changed activity, e.g. because they have the same key regulators. Moreover, it gives a quick overview over the relationships between the different types of functional sets (e.g. particular Gene Ontology biological process categories may be very similar to particular pathways contained in pathway databases). Last but not least, the color coding of edges can provide clues about potentially dysregulated crosstalks between different biological processes. The overlap graph visualization environment features a filter that can be applied to edges in order to highlight only the closest relationships between functional gene/metabolite sets.
Figure 2.
Functional gene set overlap graph summarizing predefined gene sets (and their pairwise overlaps) that are over-represented in an input list of 410 genes differentially expressed after treatment of human hepatocite-like cells with the genotoxic chemical benzo[a]pyrene. Benzo[a]pyrene causes mutations in the DNA and leads to carcinogenesis (38). Each node in the overlap graph is a predefined gene set (blue label: curated pathway, purple label: Gene Ontology category, green label: NEST and orange label: protein complex). The node size reflects the size of the gene set and the node color—its P-value (deeper red means smaller P-value). Each edge denotes an overlap between gene sets (i.e. shared genes). The edge width reflects the size of the overlap and its color reflects the number of genes/metabolites from the input list that are contained in the overlap. Details are shown in tooltips.
To exemplify how this overlap graph feature of ConsensusPathDB can be used for interpreting transcriptomics/proteomics data, Figure 2 displays results from a toxicogenomics context. Here, functional gene sets are shown that are significantly over-represented (P < 0.05) in an input list of 410 genes that appeared differentially expressed (P < 0.01) in an in vitro assay of human hepatocite-like cells that were treated with the genotoxic chemical benzo[a]pyrene, compared with an untreated control (38). The functional gene sets include manually curated pathways, Gene Ontology categories, NESTs and protein complexes that overlap with each other in different extent. The largest module of overlapping functional sets visible in Figure 2 is formed by genotoxic stress response pathways related with p53, DNA damage, apoptosis and cancer signaling. The module also includes gene sets centered at several ubiquitin E3 ligases: COP1 [gene symbol: RFWD2, a negative regulator of p53 (39)], RBX1 (Gene Ontology annotation: DNA repair) and DDB2 complex [involved in DNA repair (40)]. The results are in line with the fact that benzo[a]pyrene is a highly carcinogenic compound due to its mutagenic nature. Confirmatory, the benzo[a]pyrene metabolism pathway from WikiPathways forms a separate module together with estrogen metabolism pathways from PharmGKB and WikiPathways (upper right part of Figure 2). A third module is formed by gene sets associated with the mitochondrial ribosome (upper left part in Figure 2).
Gene set analysis based on protein complexes
A further new feature of the ConsensusPathDB web interface is the over-representation/enrichment analysis of gene lists based on curated protein complexes [in addition to functional gene sets defined over curated pathways, Gene Ontology categories and NESTs, as reported previously (8)]. ConsensusPathDB currently contains 12 263 unique curated protein complexes originating from overall 10 resources. Totally 4070 complexes have at least four distinct protein components and thus define functional sets whose size (i.e. number of member genes) is adequate for statistical over-representation/enrichment tests. These 4070 protein complex-based functional gene sets contain a total of 4645 unique genes. Notably, many of these gene sets do not correspond to any human-curated pathways or otherwise defined gene categories.
Induced network modules analysis with gene lists
In addition to the over-representation/enrichment analysis of predefined functional gene sets as detailed above, the web interface of ConsensusPathDB now provides another approach for the interaction- and pathway-centric analysis of lists of genes, called induced network modules analysis (41). Given a list of so-called seed genes (e.g. resulting from microarray experiments, which are unable to directly disclose the functional relationships between genes), it aims to interconnect those genes through different types of interactions (e.g. physical, biochemical, regulatory, etc.; selectable by the user) (Figure 3). This information on the pairwise functional/physical relationships between the genes can shed light on the biological reasons why they are identified together in the experiment. For example, if a group of genes found to be over-expressed in a microarray experiment are highly interconnected through physical interactions, this suggests that those genes may encode proteins which together form a protein complex that has a high concentration in the phenotype under study and thus may be relevant for this phenotype.
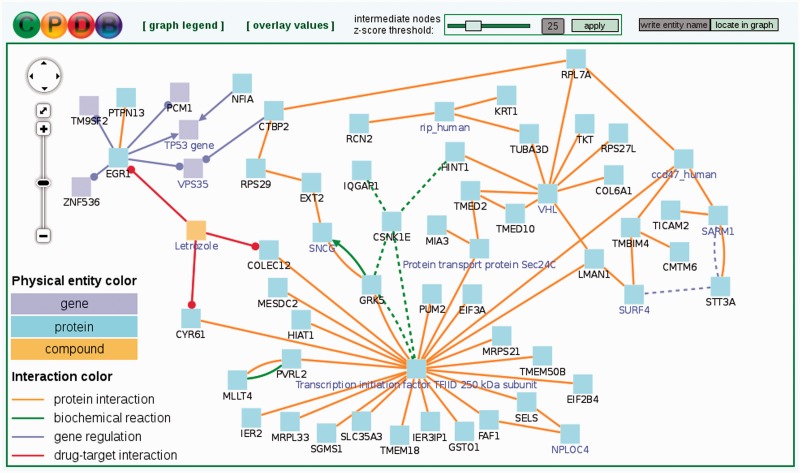
Figure 3.
Induced network module analysis of a cancer-related gene list. Each node represents a physical entity (gene, protein or compound). Nodes with black labels are from the input gene list (seed nodes) and nodes with a purple label are intermediate nodes that are not in the input list but connect seed nodes and have significantly many links in the induced network module. Each edge represents an interaction (physical, biochemical, regulatory or drug–target interaction). Numerical values can be overlaid on nodes (Supplementary Figure S1). This example network resulted from an induced network module analysis of 100 genes differentially expressed in metastatic prostate cancer as compared to non-metastatic primary prostate carcinoma and may represent a module that governs the metastatic potential of prostate cancer.
Notably, the induced network modules may optionally include genes that are not in the user-supplied seeds list, but associate two or more seed genes with each other and overall have significantly many connections within the induced network module (Figure 3, nodes with purple labels). These so-called intermediate genes could be associated with the phenotype under study, although they may not be regulated on the transcriptional level and thus do not appear in the input gene list. For example, if a group of seed genes are all connected through gene regulatory interactions to an intermediate node that represents a transcription factor, this suggests that the transcription factor may be dysfunctional (e.g. due to a mutation, which does not necessarily impact the transcription factor’s expression). Intermediate genes are ranked according to the significance of association with the seeds list given their overall connectivity in the background network. This is quantified by a z-score calculated for each intermediate node with the binomial proportions test as per Berger et al. (41). The z-score threshold can be controlled dynamically by the user in order to create sub-networks involving many intermediate and seed genes with a less stringent threshold or more compact sub-networks with a more stringent threshold.
Berger et al. (41) originally suggested the induced network modules approach and implemented it as a web tool called Genes2Networks. Their tool is limited to physical protein–protein interactions only that furthermore originate from a much smaller number of sources compared with ConsensusPathDB. Nevertheless, Genes2Networks allows the user to replace the default background network by a custom one, if available. The induced network modules analysis of ConsensusPathDB additionally features the possibility to overlay user-specified numerical values (e.g. expression values or fold changes) on nodes (genes/proteins) of the visualized network. Upon upload of a two-column, tab-delimited file containing gene accession numbers in the first column and numerical values in the second column, the values are encoded in the node color (green denoting low, negative values and red denoting high, positive values) to facilitate their visual interpretation in the context of the network (Supplementary Figure S1). The values may even be artificially created to reflect groupings of genes/proteins, e.g. according to their sub-cellular localization.
Figure 3 depicts a network module induced by a list of genes differentially expressed in metastatic prostate cancer compared with primary prostate carcinoma [data obtained from (42) and available as an example gene set on the ConsensusPathDB web site]. The module is held together by different types of interactions, comprising protein–protein, biochemical, gene regulatory and drug–target interactions. Many intermediate nodes (Figure 3, nodes with purple labels) are known cancer-associated genes although, per definition, they are not present in the input set of genes differentially expressed in metastatic prostate cancer. Examples include TP53, TAF1 (node name: ‘Transcription initiation factor TFIID 250 kDa subunit’), VHL and SNCG. Interestingly, the breast cancer drug letrozole is also present in the module and connects the seed genes EGR1, CYR61 and COLEC12 through drug–target interactions. Furthermore, the induced network modules analysis suggests metastatic prostate cancer association of RPAIN (node name: ‘rip_human’; Gene Ontology annotation: DNA repair), VPS35 (Gene Ontology annotation: cell death) and a few other genes that appear as intermediate nodes. Overall, the module constitutes an interaction network ‘cold-spot’, since most of its members are under-expressed (Supplementary Figure S1).
Other improvements
Graph visualization tool
The graph visualization tool of ConsensusPathDB was re-implemented using state-of-the-art web technology: the new visualization facilities are based on Cytoscape.js (https://github.com/cytoscape/cytoscape.js), a recently developed open-source graph visualization library for web applications, which is written in JavaScript/HTML5 as a jQuery (http://jquery.com/) plugin. The older visualization tools are deprecated since they allowed less flexibility; the old Java-based tool additionally relied on Java Virtual Machine installation.
BioPAX Level 3 export
Networks viewed with the interaction visualization tool can now be exported in BioPAX Level 3 (4) format. This format is more descriptive than previous levels and allows a more precise standard description of the sub-network of interest. For example, BioPAX Level 3 is able to represent genetic and gene regulatory interactions, which was not possible in BioPAX Levels 2 or 1.
Display of drug information for genes/proteins
The integration of drug–target interactions with physiological ones (biochemical reactions, physical interactions, etc.) mentioned above is advantageous when it comes to interaction graph-centric analysis of disease phenotypes. For example, we have previously described a new class of functional gene sets called NESTs (8). A NEST is a set of genes that are linked through different types of interactions (possibly originating from different interaction databases) to a certain gene, which is itself also included in the NEST. Given an interaction network of genes, each gene and its direct network neighbors define a separate NEST. We have shown that NEST analysis in the context of gene expression data can aid the identification of disease-causing genes (8). If available, drug information is now shown for every gene/protein in the web interface of ConsensusPathDB (including the visualization tool). Thus, ConsensusPathDB can now serve for identifying a potential target for pharmaceutical treatment and, at the same time, for retrieving information on available drugs for that target.
Improvements of the ConsensusPathDB web services
We have extended the functionality of the ConsensusPathDB web services by adding enrichment analysis functions for lists of genes or metabolites. The repertoire of predefined gene sets that can be analyzed through gene set over-representation or enrichment analysis has been extended to include NESTs, Gene Ontology categories and protein complexes in addition to curated pathways. Thus, the web services now cover completely the gene/metabolite set over-representation/enrichment analysis functionality of the web interface.
CONCLUSION
Through the integration of 30 public interaction/pathway resources, ConsensusPathDB assembles to our knowledge the most comprehensive available map of human interactions and pathways. With regular content updates and database releases every 3 months, it is ensured that this map stays up-to-date. New databases are integrated into ConsensusPathDB at the rate of 1–2 databases per release; furthermore, new interaction types are occasionally added. The recent extensions of the web interface functionality, most of which serve for the interaction- and pathway-based interpretation of sets of genes coming e.g. from transcriptomics/proteomics studies, sets of metabolites e.g. from metabolomics measurements, and the integration of drug data with physiological interactions, open further perspectives for ConsensusPathDB applications in systems biomedicine and translational research.
AVAILABILITY
The web interface of ConsensusPathDB is freely available to academic users at http://ConsensusPathDB.org. Information on web service access is provided on the ConsensusPathDB web page. The protein interaction part of the database content is available for download in tab-delimited and PSI-MI 2.5 formats on the web site. The gene compositions of biochemical pathways contained in ConsensusPathDB are available for download on the web site in a gene identifier namespace selectable by the user. Custom networks constructed by the user through interaction searches are downloadable in BioPAX Level 3 format. ConsensusPathDB can also be used for evidence mining of user-specified protein–protein interactions (e.g. obtained from an interaction screen) through a Cytoscape plugin (43). Moreover, separate ConsensusPadthDB instances exist for the model organisms, mouse (http://ConsensusPathDB.org/MCPDB) and yeast (http://ConsensusPathDB.org/YCPDB).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1 and Supplementary Methods.
FUNDING
The European Commission’s Seventh Framework Programme [DiXa, 283775]; German Ministry of Education and Research [MedSys PREDICT, 0315428A; NGFNp, NeuroNet-TP3, 01GS08171]; Max Planck Society. Funding for open access charge: European Commission.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to the developers of all source databases who have provided interaction data to the public domain. We would also like to thank the ConsensusPathDB users who have provided valuable feedback. ConsensusPathDB is developed exclusively with open-source software whose contributors are gratefully acknowledged.
REFERENCES
- 1.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 2.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, et al. The HUPO PSI’s molecular interaction format–a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 4.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D’Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aranda B, Blankenburg H, Kerrien S, Brinkman FSL, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark C, Breitkreutz B-J, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isserlin R, El-Badrawi RA, Bader GD. The Biomolecular Interaction Network Database in PSI-MI 2.5. Database. 2011 doi: 10.1093/database/baq037. January 12 (doi: 10.1093/database/baq037; epub of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan THW, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chautard E, Fatoux-Ardore M, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011;39:D235–D240. doi: 10.1093/nar/gkq830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beuming T, Skrabanek L, Niv MY, Mukherjee P, Weinstein H. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics. 2005;21:827–828. doi: 10.1093/bioinformatics/bti098. [DOI] [PubMed] [Google Scholar]
- 15.Yang C-Y, Chang C-H, Yu Y-L, Lin T-CE, Lee S-A, Yen C-C, Yang J-M, Lai J-M, Hong Y-R, Tseng T-L, et al. PhosphoPOINT: a comprehensive human kinase interactome and phospho-protein database. Bioinformatics. 2008;24:i14–i20. doi: 10.1093/bioinformatics/btn297. [DOI] [PubMed] [Google Scholar]
- 16.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luc P-V, Tempst P. PINdb: a database of nuclear protein complexes from human and yeast. Bioinformatics. 2004;20:1413–1415. doi: 10.1093/bioinformatics/bth114. [DOI] [PubMed] [Google Scholar]
- 18.Korcsmáros T, Farkas IJ, Szalay MS, Rovó P, Fazekas D, Spiró Z, Böde C, Lenti K, Vellai T, Csermely P. Uniformly curated signaling pathways reveal tissue-specific cross-talks and support drug target discovery. Bioinformatics. 2010;26:2042–2050. doi: 10.1093/bioinformatics/btq310. [DOI] [PubMed] [Google Scholar]
- 19.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F, Zhang L, Song Y, Liu X, Zhang J, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11:501–505. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Elefsinioti A, Ackermann M, Beyer A. Accounting for redundancy when integrating gene interaction databases. PLoS One. 2009;4:e7492. doi: 10.1371/journal.pone.0007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis A-R, Simonis N, Rual J-F, Borick H, Braun P, Dreze M, et al. Literature-curated protein interaction datasets. Nat. Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy ED, Landry CR, Michnick SW. How perfect can protein interactomes be? Sci. Signal. 2009;2:pe11. doi: 10.1126/scisignal.260pe11. [DOI] [PubMed] [Google Scholar]
- 28.Kamburov A, Grossmann A, Herwig R, Stelzl U. Cluster-based assessment of protein-protein interaction confidence. BMC Bioinformatics. 2012;13:262. doi: 10.1186/1471-2105-13-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamburov A, Stelzl U, Herwig R. IntScore: a web tool for confidence scoring of biological interactions. Nucleic Acids Res. 2012;40:W140–W146. doi: 10.1093/nar/gks492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis RK, Oresic M, Vidal-Puig A. Pathways to the analysis of microarray data. Trends Biotechnol. 2005;23:429–435. doi: 10.1016/j.tibtech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38:W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chagoyen M, Pazos F. MBRole: enrichment analysis of metabolomic data. Bioinformatics. 2011;27:730–731. doi: 10.1093/bioinformatics/btr001. [DOI] [PubMed] [Google Scholar]
- 34.Kamburov A, Cavill R, Ebbels TMD, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- 35.Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fowlkes EB, Mallows CL. A method for comparing two hierarchical clusterings. J. Am. Statist. Assoc. 1983;78:553–569. [Google Scholar]
- 38.Yildirimman R, Brolén G, Vilardell M, Eriksson G, Synnergren J, Gmuender H, Kamburov A, Ingelman-Sundberg M, Castell J, Lahoz A, et al. Human embryonic stem cell derived hepatocyte-like cells as a tool for in vitro hazard assessment of chemical carcinogenicity. Toxicol. Sci. 2011;124:278–290. doi: 10.1093/toxsci/kfr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 40.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 41.Berger SI, Posner JM, Ma’ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 43.Pentchev K, Ono K, Herwig R, Ideker T, Kamburov A. Evidence mining and novelty assessment of protein-protein interactions with the ConsensusPathDB plugin for Cytoscape. Bioinformatics. 2010;26:2796–2797. doi: 10.1093/bioinformatics/btq522. [DOI] [PMC free article] [PubMed] [Google Scholar]