Abstract
We present here MetalPDB (freely accessible at http://metalweb.cerm.unifi.it), a novel resource aimed at conveying the information available on the three-dimensional (3D) structures of metal-binding biological macromolecules in a consistent and effective manner. This is achieved through the systematic and automated representation of metal-binding sites in proteins and nucleic acids by way of Minimal Functional Sites (MFSs). MFSs are 3D templates that describe the local environment around the metal(s) independently of the larger context of the macromolecular structure embedding the site(s), and are the central objects of MetalPDB design. MFSs are grouped into equistructural (broadly defined as sites found in corresponding positions in similar structures) and equivalent sites (equistructural sites that contain the same metals), allowing users to easily analyse similarities and variations in metal–macromolecule interactions, and to link them to functional information. The web interface of MetalPDB allows access to a comprehensive overview of metal-containing biological structures, providing a basis to investigate the basic principles governing the properties of these systems. MetalPDB is updated monthly in an automated manner.
INTRODUCTION
It has been estimated that 30–40% of proteins require one or more metal ions to be able to carry out their biological function in cells (1,2). This proportion depends on the specific organism or tissue under consideration, which affects also the relative usage of the various metals. Additionally, metal ions play a decisive role in stabilizing the structure of nucleic acids (3). The analysis and comprehension of the interaction of metals with biological macromolecules is thus an important aspect of structural biology, e.g. to fully understand the mechanistic aspects of catalysis by metalloenzymes.
The above considerations have fostered the development of various databases aimed at providing users with a deeper insight in the three-dimensional (3D) structure of the adducts between biological macromolecules and metal ions or metal-containing cofactors (4–6). Such structures are available from the Protein Data Bank (PDB, http://www.rcsb.org/pdb/) (7). The detailed features of metal ion coordination have received particular attention in these studies. Metal ion coordination is certainly where the analysis of a metal-containing biomacromolecular structure should start from. However, the determination of such a structure is typically aimed at improving our knowledge on the functional/biochemical relevance of the interaction between the metal ion(s) and the biological macromolecule. To truly fulfil this goal, one has to go beyond the details of metal coordination, albeit these are very important (8,9). Along this line of thinking, some of us have recently demonstrated that the comparative analysis of the local structural features around the metallic cofactor and its ligands are extremely informative on the functional role of the metal itself (10,11). In practice, we defined a Minimal Functional Site (MFS) in a metalloprotein as the ensemble of atoms containing the metal ion or cofactor, its ligands and any other atom belonging to a chemical species within 5 Å from a ligand. The MFS describes the local 3D environment around the cofactor, independently of the larger context of the protein fold in which it is embedded. The systematic structural comparison of MFSs of zinc proteins allowed a structure-based classification to be developed that is tightly connected to the functional properties of each site (10). Consequently, this classification is potentially useful to predict function from 3D structure in the absence of experimental biochemical data.
The MFS concept outlined above has its chemico-physical foundation in the fact that the local environment of the metal has a determinant role in tuning its properties and thus its chemical reactivity, whereas the rest of the protein matrix is instrumental to determine, e.g. substrate selection (12) or partner recognition (13). The detailed analysis of the MFS(s) in a metal-containing biomacromolecule should constitute an important dimension of the future development of the study of metals in biology, at least in what regards its 3D structural aspects. To allow the scientific community to exploit this newly introduced perspective and simultaneously effectively and easily leverage on the vast amount of structural information that is already available, we have investigated the occurrence of MFSs in the entire PDB database and stored the corresponding results in a publicly accessible database, called MetalPDB, which is described in the present contribution. For metalloproteins, we provide an extensive set of additional contents specifically geared toward the provision of information supporting and enhancing functional interpretation. This concept is reflected in both the underlying design of the database and the way the results are presented.
DATABASE CONSTRUCTION
The construction of the MetalPDB database took place through the following steps:
Download the coordinates for all structures in the PDB.
Process each coordinate file to identify all metal atoms in the structure.
For each metal atom in each structure from step (2) identify the ligands to it. Ligands are chemical species that contain at least one non-hydrogen atom at a distance smaller than 3.0 Å from the metal. They can be residues in a polypeptide or a polynucleotide chain (endogenous ligands) as well as different ions or molecules such as water, sulfide, acetate (exogenous ligands). Organic cofactors such as heme are considered exogenous ligands.
Each pair of metal atoms that have at least one common ligand, such as a bridging amino acidic side chain or exogenous anion, or whose distance is lower than 5 Å is included into a single polynuclear site. This procedure is iterated such that if metal A and metal B are to be included into a single site and then metal B and metal C are also to be included in a single site, eventually a three-nuclear site is formed that contains all three metal ions. This procedure allowed us to define, e.g., each Fe4S4 cluster found in ferredoxins as an individual four-nuclear site.
Identify the neighbors of all the ligands (both endogenous and exogenous) to the metal atom(s) in each mono- or polynuclear site. Ligand neighbors are chemical species (residues in a polypeptide or a polynucleotide chain, or other molecules or ions) that contain at least one non-hydrogen atom at a distance smaller than 5.0 Å from the ligand itself. The ensemble of the neighbors, the ligands and the metal atom(s) constitute the MFS. H-bond interactions between ligands and ligand neighbors are identified using the HBPLUS program (14).
For each protein chain in a PDB structure, identify the 50% sequence identity group in the PDB, the EC number, if relevant, as well as the UniProt (http://www.uniprot.org/) (15), CATH (http://www.cathdb.info/) (16), SCOP (http://scop.mrc-lmb.cam.ac.uk/scop/) (17) and Pfam (http://pfam.sanger.ac.uk/) (18) codes. Each MFS is then associated with the CATH, SCOP and Pfam code(s) of the protein domain(s) that contain the ligands.
Group MFSs into sets of ‘equivalent’ and ‘equistructural’ MFSs. Two MFSs are defined to be ‘equivalent’ when they satisfy the following conditions: (i) they have the same CATH, SCOP or Pfam classification; alternatively, the sequence identity between the two PDB chains that contain them is ≥50% (effectively meaning that the two chains have the same fold (19)); (ii) after structural superposition of the PDB chains containing them, the two MFSs are superimposed (i.e. the distance between their geometric centers is <3.5 Å); and (iii) after structural superposition of the PDB chains containing them, the two MFSs have the same metal elements in the same positions. For the latter condition to be fulfilled, equivalent sites must have the same nuclearity. Two MFSs are defined to be ‘equistructural’ when they satisfy conditions (i) and (ii) above, while condition (iii) does not need to be fulfilled. This implies that two equivalent sites are also equistructural, but the converse is not necessarily true. All equivalent and equistructural MFSs are grouped into clusters of equivalent and equistructural MFSs, respectively, by using a single linkage clustering strategy. For each group of equivalent MFSs, a representative MFS is chosen by selecting the PDB structure with the highest resolution. The present step is applied to metalloproteins only as CATH, SCOP and Pfam classifications are not available for nucleic acids. Hence, no equivalent or equistructural site is defined for nucleic acids.
The steps described above are performed using python code written in house, which makes also use of the p3d module (20).
To update the database the above-mentioned procedure is identically repeated, with the only exception of restricting the query at step 1 to structures released/updated in the PDB since the latest update of MetalPDB. The update status of MetalPDB can be found in the Statistics page. At the time of submission, the database contains 175 115 sites extracted from 31 185 PDB structures out of a total of 85 045 (i.e. 36.7% of all PDB structures contained at least one metal). Fifty-five metal elements are present in at least one site in MetalPDB. The average (3272 sites) and median (118 sites) values for the number of sites, computed excluding metals with no sites, identified per metal element are very unlike. Indeed, MetalPDB includes metals for which a very large number of sites exist (magnesium being the most significant, with over 83 000 sites), next to metals with a very small number, down to only one (indium), of sites. Mononuclear sites are largely predominant (149 476, i.e. 85%) with respect to polynuclear sites (25 639, i.e. 15%).
MetalPDB can be accessed and queried via the web at the address http://metalweb.cerm.unifi.it. The user interface of MetalPDB is based on Pylons, which is an open source web application framework written in Python. Since Pylons has no default database library we chose an external database access suite, SQLAlchemy, to manage the information in the database. SQLAlchemy is an open source SQL statement builder and object-relational mapper. The database is based on PostgreSQL, which is an object-relational database system.
BROWSING METALPDB
As already mentioned, MetalPDB can be queried using the web interface at http://metalweb.cerm.unifi.it/. The interface offers via the Search menu various options to interrogate the database:
By PDB code (this is the default query form presented to the user, and is also present in the Keyword search sub-menu);
By EC Number (in the Keyword search sub-menu);
By Macromolecule name (in the Keyword search sub-menu);
By UniProt id (in the Keyword search sub-menu);
By metal element (clicking on a periodic table image);
By performing a BLAST search (for protein sequences); and
Through an Advanced Query interface.
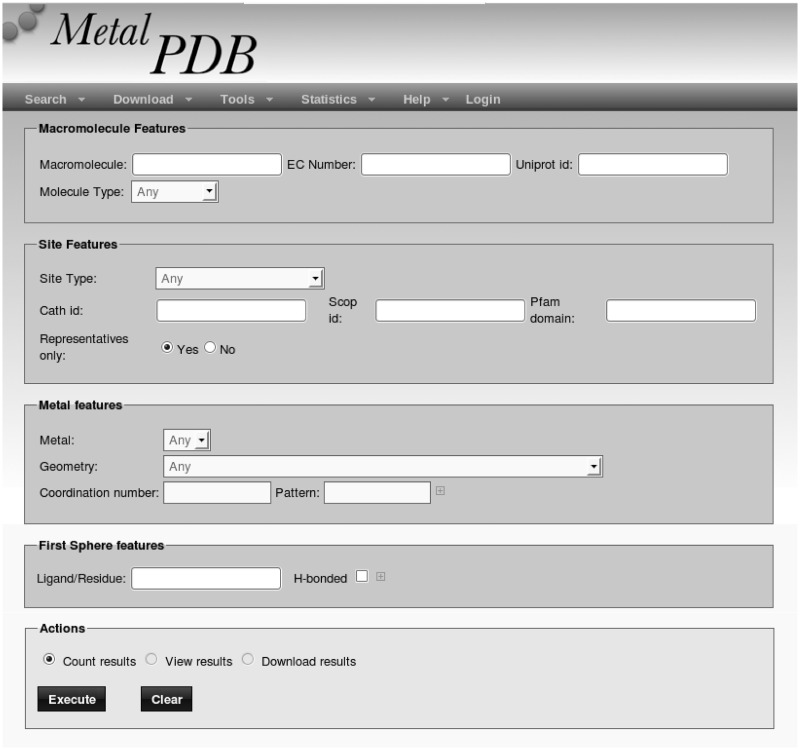
Searches may return a single database entry (e.g. when searching by PDB code) or multiple entries (e.g. sequence searches). In all cases, the user is presented with a list containing all relevant sites. The user is then asked to select one site within the list to proceed. For some queries and all advanced queries, it is possible to browse within the result list using a text box. Although the database is site-centric, the majority of the MetalPDB query types are protein-centric. A site is associated with a protein (as well as a nucleic acid) chain when that chain provides at least one metal ligand. Consequently, the database queries actually return all the sites that comprise at least one ligand belonging to the protein of interest. Sites that do not include any protein ligand, e.g. the potassium site of the 2QBY structure, are retrieved by metal searches or when searching by PDB code. In the latter case, this is possible because a site lacking any protein ligand is nevertheless associated to the PDB id of the structure containing it. Consequently, a PDB code search will retrieve more hits than, e.g., a BLAST search for the corresponding protein sequence if that structure contains one or more metal sites without protein ligands. The Advanced Query interface (Figure 1) allows users to formulate queries with varying degrees of complexity, by specifying, e.g., molecule type (protein, nucleic acid, etc.), site nuclearity, metal contents and metal geometry. The various conditions set are connected by logical AND operators. Furthermore, this interface allows the results of a query to be downloaded to a file rather than visualized (which is instead the only option available for simple queries).
Figure 1.
The Advanced Query interface of MetalPDB.
After selecting a site, the user is transported to the corresponding Summary page (Figure 2A). The Summary page displays some of the site features that have been computed at the time of its insertion in the database. The features displayed extend beyond those strictly needed for the definition of the site itself (such as nuclearity or the identity of the ligands) and include, among others, the EC number of the chain(s) containing the site (interfacial sites are associated with more than one chain), the coordination geometry of each metal in the site (computed using our FindGeo approach (21), which can be straightforwardly applied from the same portal to obtain more detailed parameters such as rms deviation from the ideal geometry), the structural or domain classification of the chain(s) containing the site. Additionally, the coordinates of the site can be downloaded or the site inspected using Jmol (22). A schematic representation of the site is also available.
Figure 2.
Overview of the results of a query to MetalPDB (using the structure 2SOD as an example). (A) The Summary page. (B) Detailed description of the first coordination sphere of the metals in the site (only copper is shown). (C) List of the representative equistructural sites.
Next to the Summary page, there are three tabs available:
Coordination sphere;
Equivalent sites; and
Equistructural sites.
The Coordination sphere tab provides more detailed information for each metal in the site on coordination as well as other structural properties (Figure 2B). Indeed, the tab contains a large table for each metal that is further subdivided to display or permit access to metal properties. For example, donor atom names, types and distances from the metal are given in tabular form. In addition, for each ligand it is possible to display and/or download tables reporting hydrogen bonding or van der Waals interactions. The same information can be schematically visualized. The rightmost column of each metal table shows a plot of the metal environment.
Under the Equivalent sites tab the user can find a list of sites that are equivalent to the site currently displayed (see the ‘Database construction’ section). Equivalent sites can be found in different PDB structures having the same fold, or in different but identically folded chains within the same PDB structure. In a nutshell, the list of equivalent sites contains all MFSs present in the PDB databank that contain the same metal in the same position as the current MFS, within a structure with the same fold as the structure containing the current site. However, the ligands may differ, although this is not common. Instead, the neighbors to the ligands will typically differ, to an extent depending largely on the sequence similarity between the protein chains compared (23). Thus, the Equivalent sites tab allows users to readily identify families of proteins containing the same MFS, facilitating them to deal with the far from trivial task of assessing the redundancy of PDB structures in terms of their metal content. The coordinates of all the superimposed sites can be immediately downloaded from MetalPDB, together with a very simple Pymol script to visualize them.
Under the Equistructural sites tab (Figure 2C) the user can find a list of sites that are equistructural to the MFS currently displayed (see the ‘Database construction’ section). As previously noted, two equistructural sites may or may not be also equivalent. For simplicity, the download button in the MetalPDB interface allows users to download a table of equistructural sites that are not equivalent to the MFS of interest (the latter can be obtained via the Equivalent sites tab). In practice, MFSs that are equistructural but not equivalent are sites in corresponding positions within protein structures having the same fold while they differ for their metal contents. This can happen for a variety of reasons. Metal ions can replace one another within the same site for both physiological and non-physiological reasons (24–26) or upon in vitro chemical treatment [typically to introduce spectroscopically active metals (27,28)]. Engineering of the metal ligands or of their neighbors can affect the relative affinity of a site toward different metal ions, eventually leading to incorporation of different metals in mutants with respect to the wild-type protein (29,30). For polynuclear sites, it is additionally possible to observe phenomena such as the incorporation of different sets of metal ions (which again can be physiologically relevant or entirely due to in vitro treatment, and can change the nuclearity of the site), replacement of some or all of the metal ions with others [e.g. as observed in phosphatases (31,32)]. Each equistructural site shown in the tab is the representative (i.e. the site in the structure with the highest resolution) of a group of equivalent MFSs: the sites equivalent to these representatives are not shown to allow users to grasp immediately the variation range independently of the number of MFSs in each group (Figure 2C).
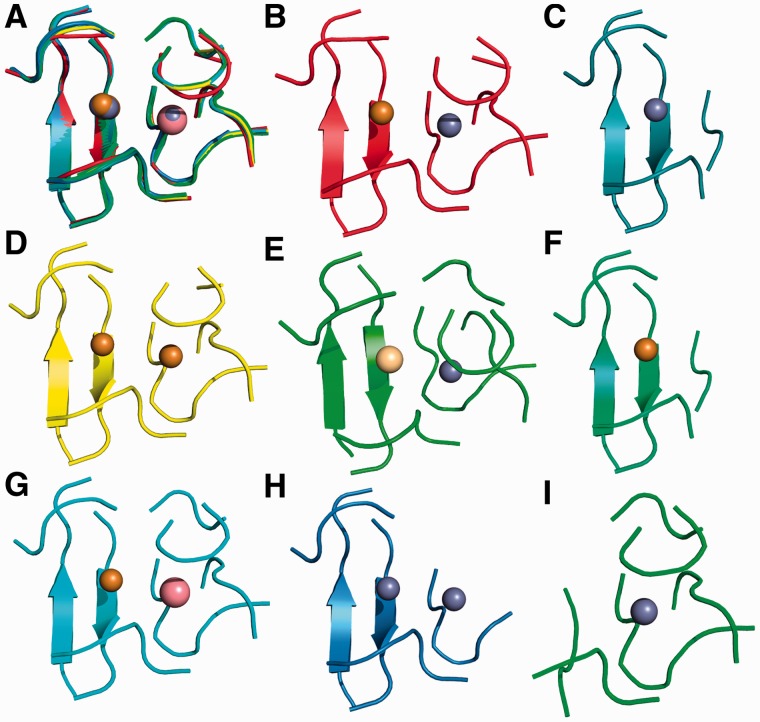
As an example, Figure 3 shows equistructural sites for human superoxide dismutase [hSOD, PDB code 2SOD (33)]. hSOD contains a dinuclear site containing one zinc ion and one copper ion. The equistructural sites differ from it in the number and/or nature of the metal ion occupying each position within the 2SOD site. In particular, the equistructural sites can be of reduced nuclearity, i.e. mononuclear, or can contain a different metal in one or both site positions (e.g. a zinc–zinc cluster). Figure 2C shows that there are two families of mononuclear equistructural sites containing one zinc ion. This can happen because the zinc ion in each of the two families occupies only one of the two positions in the dinuclear site (Figure 3C and I, respectively).
Figure 3.
Equistructural sites for hSOD (PDB code 2SOD). The site in 2SOD has eight groups of equistructural sites, including its own group, encompassing a total of 579 sites. Only one representative for each group is shown. All sites are displayed using a cartoon representation of the protein backbone, whereas the metal ions are represented as spheres. The color code for the metal ions is as follows: gray-zinc, gold-copper, pink-cobalt and light yellow-cadmium. (A) Superimposition of all sites. Each site maintains the same color as in its individual panel; (B) 2SOD; (C) 2C9S; (D) 1E9P; (E) 1MFM; (F) 1Q0E (copper site); (G) 1COB; (H) 3H2P; and (I) 1Q0E (zinc site). (C) and (F) are equistructural to the left half of the full site (see panel A), whereas (I) is equistructural to the right half of the full site.
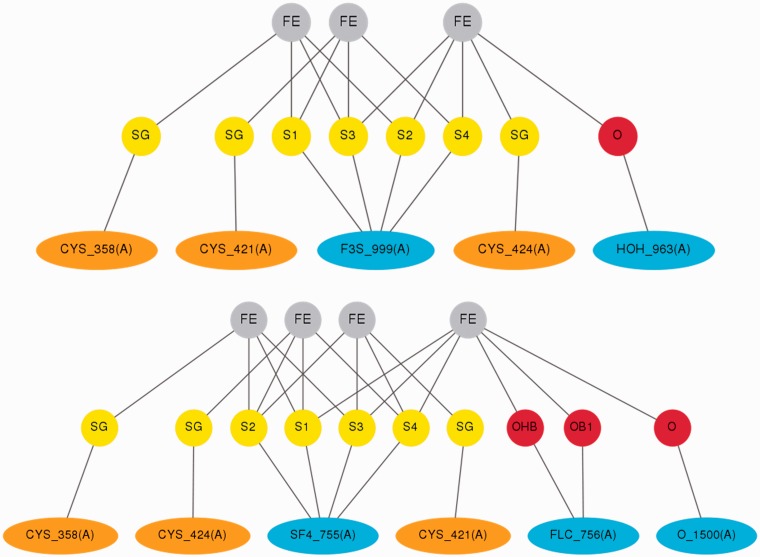
A further example of a possible use of MetalPDB is given in Figure 4. Here we exploit the site schemes that are produced by our tools and can be displayed from the Summary page (Figure 2A). The case represented is that of aconitase, EC number 4.2.1.3, whose function is sensitive to mitochondrial iron deficiency and oxidative stress via the reversible conversion of a Fe4S42+ cluster into a Fe3S4+ cluster (36). The site schemes allow the user to readily compare and localize the associated changes in the site coordination sphere (Figure 4).
Figure 4.
Schematic comparison of the aconitase sites in the two functionally relevant forms. The schemes are directly taken from the MetalPDB entries associated to PDB codes 5ACN (Fe3S4 form, top) (34) and 1C96 (Fe4S4 form, bottom) (35). Note that the cluster name changes from SF4 to F3S in accordance with PDB naming standards. Water molecule 963 is at 2.97 Å from one of the iron ions in 5ACN.
COMPARISON WITH OTHER AVAILABLE DATABASES FOCUSING ON METALS IN BIOLOGY
COMe (http://www.flymine.org/come/) (37) provides only information on the first coordination sphere of the metal center, i.e. essentially what MetalPDB is providing in the first coordination sphere tab. This database has not been updated since 2005.
The MDB (http://metallo.scripps.edu/) (4) is the first database that was created for metalloproteins and is specifically geared toward providing information useful for metalloprotein design. This results in the information provided consisting mainly of a description of the features of the metal coordination environment. This database has not been updated since 2003.
MESPEUS (http://mespeus.bch.ed.ac.uk/MESPEUS_10/) (5) is a relatively recent database, implemented in 2008, which provides extensive information on the metal coordination environment of metalloproteins in the PDB, basically providing a detailed description of all geometric features of the metal site. Crystallographic features are also described extensively, and it is possible to easily generate statistics for metals in any selected environment. Whereas MESPEUS geometric insight is far more extended than what we are providing in MetalPDB, its usefulness for functional analysis is more limited. Indeed, MESPEUS does not provide any comparison between different sites, as we instead accomplish by looking at equivalent and equistructural sites, nor it provides any analysis of protein domains. This database has not been updated since 2010.
MetLigDB (http://silver.sejong.ac.kr/MetLigDB/home.html) (38) focuses on the analysis of organic ligands binding to metalloproteins and not on metal–biomacromolecule interactions; its scope is thus widely different than MetalPDB.
MINAS (http://www.minas.uzh.ch/) (6) focuses on metal–nucleic acid interactions, and thus it does not include metalloproteins. Thus MetalPDB and MINAS can be seen as complementary, with some limited overlap. Note that of the 175 115 MFS contained in MetalPDB, 86 637 (49.5%) have at least one protein ligand and no nucleic acid ligand whereas 31 452 (18.0%) have at least one nucleic acid ligand and no protein ligand and 54 594 (31.2%) have ligands that are neither proteic or nucleic (the latter MFS’s may however interact with proteins and/or nucleic acids in their second sphere).
A general observation from the analysis of the existing databases described in the preceding paragraph is that there are no metalloprotein databases that are truly kept up to date. To avoid encountering the same problem with MetalPDB, we designed it in a way that enables automated update of its contents (at present on a monthly basis, to become more frequent in the future). In fact all the analyses carried out and described in the preceding paragraphs are entirely programmed in the code underlying the construction of the database.
CONCLUDING REMARKS
MetalPDB is an innovative resource designed for all researchers interested in the study of metals in biology. This resource capitalizes on the notion that a much deeper insight into the biological relevance of metal–biomacromolecule interactions is obtained by supplementing the detailed analysis of metal coordination, which has already been extensively addressed in the literature, with the examination of the macromolecular environment in which the metal and the ligands are embedded (represented by the MFS). MetalPDB makes this analysis systematically and readily available to the scientific community. With respect to other available resources, our focus is thus shifted from metalloprotein design or crystallographic features to the functional features of metalloproteins, also through functional and structural domain analysis. A key advantage of MetalPDB over other related databases is that its contents are generated and monthly updated in an automated manner.
Finally, MetalPDB allows users to easily identify and visualize similarities as well as variations in the way metals are coordinated by proteins by linking each entry to those bearing equivalent and equistructural sites. This functionality provides direct access to otherwise complex structural comparisons.
FUNDING
Ministero Italiano dell’Università e della Ricerca (MIUR) [RBFR08WGXT, RBRN07BMCT and 2009FAKHZT]. Funding for open access charge: MIUR [RBFR08WGXT].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the technical help of Enrico Morelli.
REFERENCES
- 1.Andreini C, Bertini I, Rosato A. Metalloproteomes: a bioinformatic approach. Acc. Chem. Res. 2009;42:1471–1479. doi: 10.1021/ar900015x. [DOI] [PubMed] [Google Scholar]
- 2.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 3.Pechlaner M, Sigel RKO. Characterization of metal ion-nucleic acid interactions in solution. Met. Ions Life Sci. 2012;10:1–42. doi: 10.1007/978-94-007-2172-2_1. [DOI] [PubMed] [Google Scholar]
- 4.Castagnetto JM, Hennessy SW, Roberts VA, Getzoff ED, Tainer JA, Piquet ME. MDB: the metalloprotein database and browser at the Scripps Research Institute. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsin K, Sheng Y, Harding MM, Taylor P, Walkinshaw MD. MESPEUS: a database of the geometry of metal sites in proteins. J. Appl. Cryst. 2008;41:963–968. [Google Scholar]
- 6.Schnabl J, Suter P, Sigel RKO. MINAS–a database of metal ions in nucleic acids. Nucleic Acids Res. 2012;40:D434–D438. doi: 10.1093/nar/gkr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harding MM, Nowicki MW, Walkinshaw MD. Metals in protein structures: a review of their principal features. Crystallogr. Rev. 2010;16:247–302. [Google Scholar]
- 9.Dudev T, Lim C. Metal binding affinity and selectivity in metalloproteins: insights from computational studies. Annu. Rev. Biophys. 2008;37:97–116. doi: 10.1146/annurev.biophys.37.032807.125811. [DOI] [PubMed] [Google Scholar]
- 10.Andreini C, Bertini I, Cavallaro G. Minimal functional sites allow a classification of zinc sites in proteins. PLoS One. 2011;10:e26325. doi: 10.1371/journal.pone.0026325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreini C, Bertini I, Cavallaro G, Najmanovich RJ, Thornton JM. Structural analysis of metal sites in proteins: non-heme iron sites as a case study. J. Mol. Biol. 2009;388:356–380. doi: 10.1016/j.jmb.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 12.Bertini I, Fragai M, Luchinat C, Melikian M, Venturi C. Characterization of the MMP-12-elastin adduct. Chem. Eur. J. 2009;15:7842–7845. doi: 10.1002/chem.200901009. [DOI] [PubMed] [Google Scholar]
- 13.Banci L, Bertini I, Calderone V, Della Malva N, Felli IC, Neri S, Pavelkova A, Rosato A. Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces. Biochem. J. 2009;422:37–42. doi: 10.1042/BJ20090422. [DOI] [PubMed] [Google Scholar]
- 14.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 15.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 20.Fufezan C, Specht M. p3d–Python module for structural bioinformatics. BMC Bioinform. 2009;10:258. doi: 10.1186/1471-2105-10-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreini C, Cavallaro G, Lorenzini S. FindGeo: a tool for determining metal coordination geometry. Bioinformatics. 2012;28:1658–1660. doi: 10.1093/bioinformatics/bts246. [DOI] [PubMed] [Google Scholar]
- 22.Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 23.Bertini I, Luchinat C, Provenzani A, Rosato A, Vasos PR. Browsing gene banks for Fe2S2 ferredoxins and structural modeling of 87 plant-type sequences: an analysis of fold and function. Prot. Struct. Funct. Genet. 2002;46:110–127. doi: 10.1002/prot.10009. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt M, Meier B, Parak F. X-ray structure of the cambialistic superoxide dismutase from propionibacterium shermanii active with Fe or Mn. JBIC. 1996;1:532–541. [Google Scholar]
- 25.Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, Cheek TR, Gray J, Banfield MJ, Dennison C, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 26.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 27.Banci L, Piccioli M. Cobalt(II) and Nickel(II) substituted proteins. In: Grant DM, Harris RK, editors. Encyclopedia of Nuclear Magnetic Resonance. 1996. pp. 1365–1373. [Google Scholar]
- 28.D'Onofrio M, Gianolio E, Ceccon A, Arena F, Zanzoni S, Fushman D, Aime S, Molinari H, Assfalg M. High relaxivity supramolecular adducts between human-liver fatty-acid-binding protein and amphiphilic Gd(III) complexes: structural basis for the design of intracellular targeting MRI probes. Chemistry. 2012;18:9919–9928. doi: 10.1002/chem.201103778. [DOI] [PubMed] [Google Scholar]
- 29.Barrick D. Depletion and replacement of protein metal ligands. Curr. Opin. Biotechnol. 1995;6:411–418. doi: 10.1016/0958-1669(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y, Yeung N, Sieracki N, Marshall NM. Design of functional metalloproteins. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swingle MR, Honkanen RE, Ciszak EM. Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5. J. Biol. Chem. 2004;279:33992–33999. doi: 10.1074/jbc.M402855200. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Tainer JA, Getzoff ED, Beem KM, Richardson JS, Richardson DC. Determination and analysis of the 2 Å structure of copper, zinc superoxide dismutase. J. Mol. Biol. 1982;160:181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- 34.Robbins AH, Stout CD. Structure of activated aconitase: formation of the [4Fe-4S] cluster in the crystal. Proc. Natl Acad. Sci. USA. 1989;86:3639–3643. doi: 10.1073/pnas.86.10.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd SJ, Lauble H, Prasad GS, Stout CD. The mechanism of aconitase: 1.8 Å resolution crystal structure of the S642a:citrate complex. Protein Sci. 1999;8:2655–2662. doi: 10.1110/ps.8.12.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasquez-Vivar J, Kalyanaraman B, Kennedy MC. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. J. Biol. Chem. 2000;275:14064–14069. doi: 10.1074/jbc.275.19.14064. [DOI] [PubMed] [Google Scholar]
- 37.Degtyarenko K, Contrino S. COMe: the ontology of bioinorganic proteins. BMC Struct. Biol. 2004;4:3. doi: 10.1186/1472-6807-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi H, Kang H, Park H. MetLigDB: a web-based database for identification of chemical groups to design metalloprotein inhibitors. J. Appl. Cryst. 2011;44:878–881. [Google Scholar]