Abstract
Similarity of gene expression across a wide range of biological conditions can be efficiently used in characterization of gene function. We have constructed a rice gene coexpression database, RiceFREND (http://ricefrend.dna.affrc.go.jp/), to identify gene modules with similar expression profiles and provide a platform for more accurate prediction of gene functions. Coexpression analysis of 27 201 genes was performed against 815 microarray data derived from expression profiling of various organs and tissues at different developmental stages, mature organs throughout the growth from transplanting until harvesting in the field and plant hormone treatment conditions, using a single microarray platform. The database is provided with two search options, namely, ‘single guide gene search’ and ‘multiple guide gene search’ to efficiently retrieve information on coexpressed genes. A user-friendly web interface facilitates visualization and interpretation of gene coexpression networks in HyperTree, Cytoscape Web and Graphviz formats. In addition, analysis tools for identification of enriched Gene Ontology terms and cis-elements provide clue for better prediction of biological functions associated with the coexpressed genes. These features allow users to clarify gene functions and gene regulatory networks that could lead to a more thorough understanding of many complex agronomic traits.
INTRODUCTION
Rice (Oryza sativa L.) has been at the forefront of plant genomics since the completion of the genome sequence of the japonica cultivar Nipponbare. The high-quality map-based sequence and complete annotation of every transcriptional unit in the genome have facilitated remarkable advancements in molecular genetics of rice as well as in many applied aspects of cereal genomics (1–4). Subsequently, elucidating the function of all predicted genes has become an ultimate goal not only for complete understanding of the biology of rice but also in developing novel strategies for crop improvement. Due to worldwide efforts in functional genomics, a wide range of resources for characterizing the function of rice genes have been developed and made available to the scientific community (5–9). However, despite these extensive efforts, more than half of about 32 000 genes that define the rice plant, many of which could be possibly involved in the expression of agronomically important traits, still remains to be fully characterized.
Gene clustering analysis, which is based on the premise that clusters of genes with similar expression patterns across several experimental conditions tend to be functionally related (10), has become a powerful tool for gene function prediction. With the rapid accumulation of microarray data in public repositories, such as NCBI’s Gene Expression Omnibus (GEO) (11,12), European Bioinformatics Institute (EBI)’s Array Express (13) and DNA Data Bank of Japan (DDBJ)’s CIBEX (14), there is also a proliferation of databases on gene expression profiling of various crops in the public domain (15–19). Similarly, an increasing number of gene coexpression databases in various plant species such as Arabidopsis and rice have also been developed (20–26). Coexpression approaches have been widely used in Arabidopsis, particularly in analysis of various biological targets such as transcription factors, enzymes in specific metabolic pathways and protein subunits (27). In rice, gene coexpression data are provided by the RiceArrayNet (25), OryzaExpress (26), ATTED-II (28) and Rice Oligonucleotide Array Database (19). RiceArrayNet was developed based on the Rice 60k Microarray data, and the rest were constructed using rice Affymetrix microarray data.
In the last 5 years, we have collected >800 microarray data from organs and tissues of rice plants at various developmental stages, mature organs throughout the entire growth in the field, cell/tissue types isolated by laser microdissection and rice plants subjected to plant hormone treatment (29,30). These expression profiles generated from the model rice cultivar Nipponbare using a single microarray system (Agilent) are available in our gene expression profile database, RiceXPro (17). Most of the data were collected from rice plants grown in the natural environment, which is much more complex than controlled laboratory conditions. This large-scale field transcriptome profiling can be used as a measure of the corresponding changes in physiological and morphological states of the plant triggered by internal or external stimuli under natural conditions. Therefore, our comprehensive collection of gene expression profiles provides a more accurate landscape of the transcriptome that could be used for understanding the function of genes based on the level, time specificity and position specificity of expression, as well as the overall similarity of gene expression of various organs and tissues in the natural field environment.
Here, we describe the Rice Functionally Related gene Expression Network Database (RiceFREND), a database for retrieving gene coexpression information across a wide range of gene expression profiles which we collected using a single microarray platform.
DATABASE COMPONENTS AND FEATURES
The overall concept of RiceFREND is to provide a platform for efficient prediction of gene functions and gene regulatory networks based on gene expression similarity. A schematic diagram of the major components and various features of the database such as search options, coexpression network viewers, analysis tools and detailed information on coexpressed genes is shown in Supplementary Figure S1.
Calculation for coexpression analysis
All gene expression data described in this database were obtained from microarray analysis using the rice 4×44K microarray RAP-DB platform (Agilent Technologies, G2519F#15241), which contains 35 760 independent probes corresponding to 27 201 annotated loci published in the Rice Annotation Project Database (RAP-DB) (4). A total of 24 data sets corresponding to 815 microarray data from various organs and tissues at different developmental stages and treatment with plant hormones, including redundant data among the data sets, were used as data source for coexpression analysis (Supplementary Table S1). All data have been deposited in GEO (11,12) and accessible through GEO series accession numbers: GSE21396, GSE21397, GSE36040, GSE36042, GSE36043, GSE36044, GSE39423, GSE39424, GSE39425, GSE39426, GSE39427, GSE39429 and GSE39432.
The expression data were obtained either by intensity-based one-color or ratio-based two-color microarray hybridization system. In the 75th percentile normalization for one-color method, the processed raw signal intensities of 35 760 probes were divided by 75th percentile value and transformed to log2 scale. The median expression value within each data set was further subtracted to obtain the relative value for each probe. Mean values were used for redundant probes assigned to the same locus. For data obtained using the two-color microarray system, the expression profile for each gene corresponds to the log2 ratio of cyanine 5 (Cy5) signal intensity for treatment to cyanine 3 (Cy3) signal intensity for mock treatment (log2 Cy5/Cy3). All data sets were combined into one gene expression matrix data for coexpression analysis among genes. The weighted Pearson’s correlation coefficients were calculated to reduce any unsuitable effects that may have resulted from sample redundancies. The mutual rank (MR) was used as an index for coexpression as described in ATTED-II (21,31).
Annotation of genes
The RAP2 representative data in general feature format derived from RAP-DB (4) were basically used as annotation of each gene. The corresponding Michigan State University Osa1 Rice Locus (MSU ID) (32) for each RAP locus ID was obtained using the ID converter tool in RAP-DB. Generic Gene Ontology (GO) annotations in RAP-DB were converted to GO slim using the map2slim.pl script (http://search.cpan.org/∼cmungall/go-perl/). A total of 1308 genes commonly described in PlnTFDB (33) and PlantTFDB (34) were classified as transcription factors. The subcellular localization sites of proteins were predicted by WoLF PSORT (35) based on the deduced amino acid sequences of each gene. The gene symbols/names of 1523 genes and functional categories of 2053 genes were obtained from Oryzabase (36) and KEGG Pathway Database (37), respectively.
Coexpression search with a single guide gene
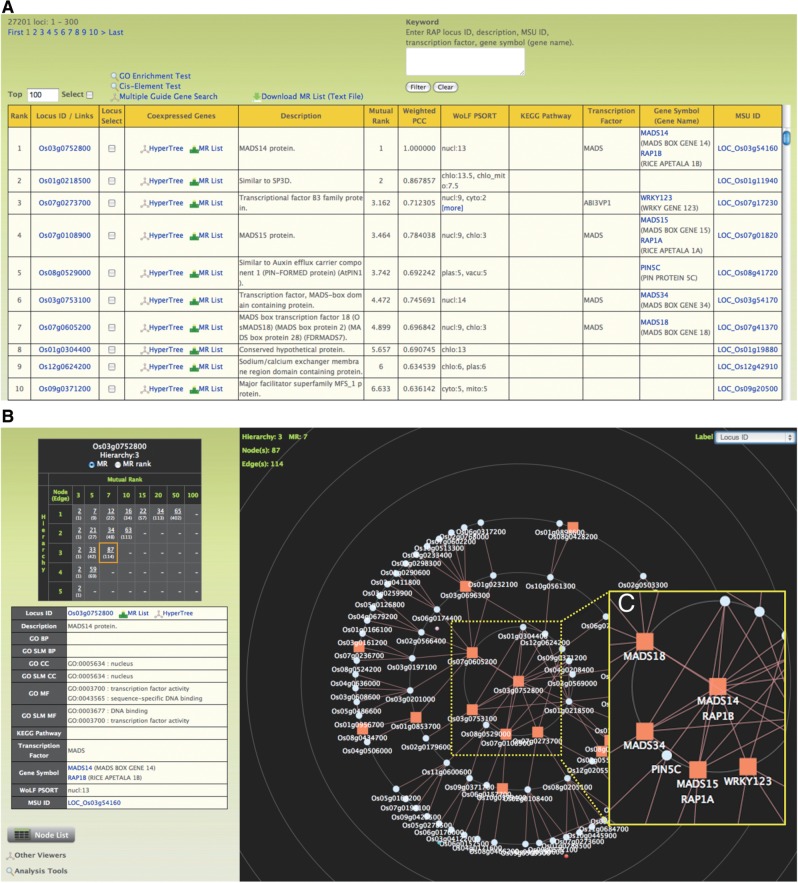
The coexpression search is based on the ‘guide gene’ approach in which a pre-selected gene or set of genes is used to retrieve coexpressed genes. In the single guide gene search option, coexpression search is initiated by entering a keyword such as RAP locus ID, gene name, MSU ID and other identifiers or by selecting specific chromosome in the panel representing the 12 rice chromosomes. This provides a tabular list of coexpressed genes with information on RAP locus ID, gene description, transcription factor, gene symbol and the corresponding MSU ID. The RAP locus ID opens a pop-up window with direct links to RiceXPro (17) for gene expression profile used in coexpression analysis, RAP-DB (4) for detailed annotations and SALAD database (38) for genome-wide comparative analysis of motifs. The MSU ID provides direct link to the annotation of the gene as described in the MSU Rice Genome Annotation Project database (32). The coexpression data for each gene are shown in two formats, a tabular list of coexpressed genes in ascending order of MR (Figure 1A) and a graphical representation of the coexpression network in HyperTree (http://philogb.github.com/jit/) format (Figure 1B). The MR list also contains information on predicted subcellular localization sites of proteins, KEGG pathways and transcription factors. Coexpressed genes can be selected and used for further analysis, such as GO enrichment test and cis-element analysis as described below. A filter option allows users to retrieve information for gene/genes of interest using RAP locus ID, description, MSU ID, transcription factor and gene symbol/name as keywords.
Figure 1.
Single guide gene search results interfaces. (A) Tabular list of coexpressed genes generated from the search with Os03g0752800 (MADS14) as guide gene. The coexpressed genes are arranged in the order of decreasing MR value. (B) Graphical view of the coexpressed gene network in HyperTree format. Two nodes representing coexpressed genes with MR value below a defined threshold are joined to form an edge. The gene network (MR <7, hierarchy = 3) shown here contains 87 nodes and 114 edges. (C) An enlarged view of the coexpression network using gene symbol as label for nodes. MADS14 is strongly coexpressed with MADS15, MADS18 and MADS34.
The HyperTree graphical viewer shows the relationships of coexpressed genes based on a user-defined hierarchy and MR value or rank based on ascending MR value. In this viewer, the nodes representing the coexpressed genes with MR value below a defined threshold are joined to form edges (Figure 1B and C). The coexpression data can be viewed for a maximum of 100 coexpressed genes and the network size can be set by selecting the MR value and hierarchy. Detailed information on the nodes comprising a network is provided from the ‘Node List’ and via a pop-up window on each node in the HyperTree. The nodes are labeled with RAP locus ID but options for other identifiers such as gene symbol, gene name, transcription factor, MSU ID and KEGG pathway are available. The node for a transcription factor is indicated by a red square and the node with a KEGG pathway annotation is indicated by a tag corresponding to a specific pathway. All genes with KEGG pathway annotation are listed in a table with direct links to the database. In addition to HyperTree, the gene network can also be viewed in Cytoscape Web (39) and Graphviz (http://www.graphviz.org/) formats, thereby providing several options to visually understand the relationship among coexpressed genes. Download options are available for the node list, data for construction of network in Cytoscape (40) and the graph images in Cytoscape Web and Graphviz formats.
The coexpressed gene network generated using MADS14 (Os03g0752800), which encode APETALA1/FRUITFUL-like MADS domain protein, as a guide gene is shown in Figure 1B and C. MADS14 is coexpressed with other MADS-box genes, namely, MADS15 (Os07g0108900), MADS18 (Os07g0605200) and MADS34 (Os03g0753100). It has been reported recently that the four MADS-box genes were coordinately expressed in the shoot apical meristem to specify the identity of the inflorescence meristem during reproductive phase transition (41). Therefore, the gene network would reflect overlapping functions of the four MADS-box genes and provided useful clue that could be used to further elucidate the molecular mechanism of reproductive transition in rice.
Coexpression search with multiple guide genes
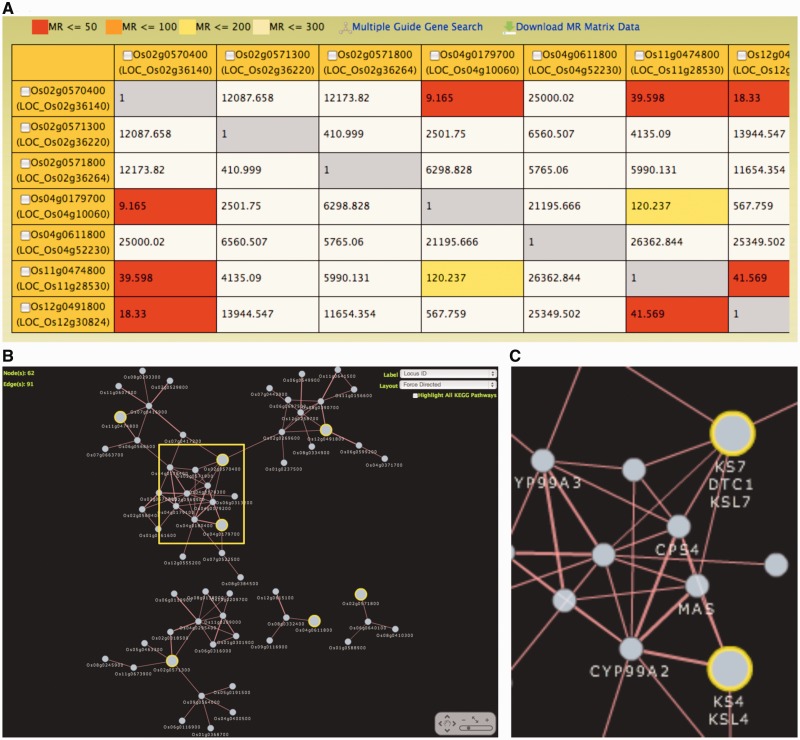
The multiple guide gene search uses two or more pre-selected genes as guide genes to expand the coverage of gene coexpression networks. This search is also initiated by entering a keyword such as RAP locus ID, gene name, MSU ID or other identifiers. From the tabular list generated by the search, two or more guide genes are selected, and the hierarchy and MR value or rank to be used for construction of gene network are defined. Information useful for selection of threshold in MR value among query genes can be viewed in the matrix data (Figure 2A). The coexpression network is generated in either a Cytoscape Web format with several options for node label and network layout (Figure 2B) or Graphviz format, both of which provide detailed annotation of nodes, direct links to other databases and the HyperTree and MR List from the single guide gene search against each node in the network. Additionally, the coexpression network can be reconfigured by selecting other guide genes from the node list panel. As in the single guide gene search, download options are also available for note list, data for construction of Cytoscape and the graph images in Cytoscape Web and Graphviz formats.
Figure 2.
Multiple guide gene search results interfaces. (A) Matrix data of MR values among multiple guide genes. Two or more guide genes can be selected to initiate coexpression search. (B) Graphical view of the coexpressed gene network in Cytoscape Web format. The guide genes are shown as large circles with yellow outline. (C) An enlarged view of the coexpression network using gene symbol as label for nodes. Genes involved in momilactone biosynthesis such as CPS4, KS4, MAS, CYP99A2 and CYP99A3 are strongly coexpressed.
The gene network generated by the multiple guide gene search (MR < 7, hierarchy = 2) with ent-kaurene synthase gene Os04g0611800 (OsKS1) and six kaurene synthase-like genes, namely, Os04g0179700 (OsKSL4), Os02g0571300 (OsKSL5), Os02g0571800 (OsKSL6), Os02g0570400 (OsKSL7), Os11g0474800 (OsKSL8) and Os12g0491800 (OsKSL10) as guide genes is shown in Figure 2B. OsKS1 is associated with gibberellin biosynthesis (42), whereas OsKSL4, OsKSL8, OsKSL10 and OsKSL7 are related to biosynthesis of diterpenoid phytoalexin such as momilactones A and B, oryzalexins S, oryzalexins A–F and phytocassanes A–E, respectively (43–45). This multiple guide gene search generated a gene network comprising four clusters. The largest cluster contains OsKSL4, OsKSL7, OsKSL8 and OsKSL10 which have been reported to be induced by UV irradiation and elicitor treatment (42–45). On the other hand, OsKS1, and OsKSL5 and OsKSL6, the expression of which is not affected by such treatment (46), were included in a separate cluster. Interestingly, momilactone biosynthetic genes (47), namely, CPS4, KSL4, MAS, CYP99A2 and CYP99A3 are strongly connected to each other (Figure 2C), and several phytocassane biosynthetic genes (48) are also included in the largest cluster. This coexpression analysis suggests that the resulting gene network can be used to define the biological function of each kaurene synthase genes and may therefore help in exploring novel enzymes related to phytoalexine biosynthesis based on the coexpression signature.
Analysis tools
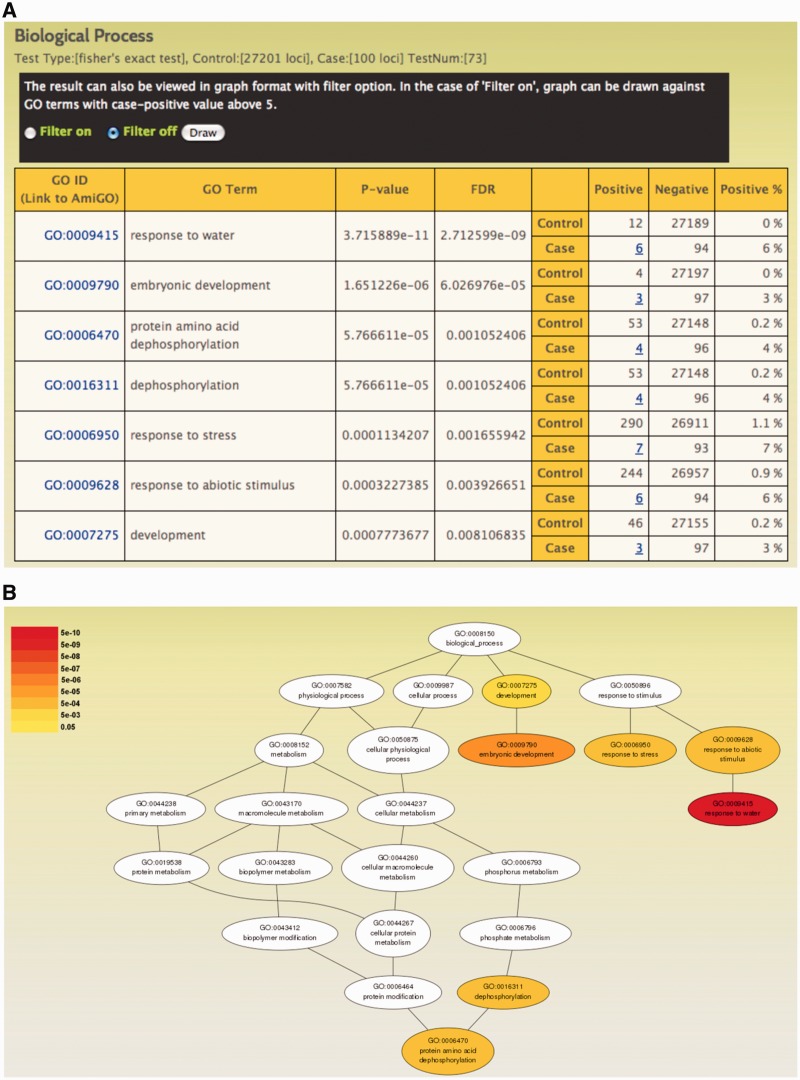
In order to enhance prediction of biological functions associated with the coexpressed gene network, RiceFREND is provided with analysis tools for identification of enriched GO terms and cis-elements. The GO enrichment test facilitates the identification of overrepresented GO terms among coexpressed genes for better understanding of the functional relationships of these genes. The Fisher’s exact test is applied against GO terms categorized into biological process and molecular function. This generates a tabular list of overrepresented GO terms in ascending order of P-value which is adjusted by the Benjamini and Hochberg’s method (49) for multiple testing collection (Figure 3A). The result can also be viewed in graph format with a filter option (Figure 3B) that can be used to construct a graph against GO terms with case-positive value >5.
Figure 3.
GO enrichment test results interfaces. (A) A tabular list of over-represented GO terms (false discovery rate, FDR < 0.05) generated by GO enrichment test against the top 100 genes coexpressed with LEA3-1. (B) The over-represented terms can be visualized in a graphical format with the enriched GO terms (FDR < 0.05) shown in color scale corresponding to the FDR value. The graph was constructed with filter off setting.
Identification of cis-elements among coexpressed genes could provide additional information for prediction of function. The cis-element analysis tool can be used to clarify whether a specific motif is overrepresented in the 1-kb upstream regions of coexpressed genes based on three statistical tests, namely, χ2 test, Fisher’s exact test and G-test.
We applied the GO enrichment test and cis-element analysis on the top 100 genes coexpressed with LEA3-1 gene (Os05g0542500). It has been reported that over-expression of LEA3-1 improves drought resistance under the natural field conditions (50). Consistent with this observation, the GO terms such as ‘response to water’, ‘response to stress’ and ‘response to abiotic stimulus’ were found to be overrepresented among the coexpressed genes (Figure 3A and B). The expression of LEA3-1 gene is induced not only by drought stress but also by treatment with abscisic acid (ABA). In the cis-element analysis, ABA-responsive element core (ACGT) containing CGMCACGTGB motif (51) is also statistically enriched in the promoter region of the coexpressed genes. Therefore, although the microarray data used for coexpression analysis do not include abiotic stress treatments such as drought and salinity, the results of coexpression search and additional analysis can be used to understand functional relationships among stress-inducible genes based on the coexpression.
FUTURE DIRECTIONS
Coexpression analysis can be used more accurately with a more balanced distribution of expression data from various organs and tissues and a wider coverage in terms of experimental conditions. We are currently undertaking microarray analysis involving various organs and tissues at different developmental stages and rice plants subjected to biotic and abiotic stresses. These data will be added to the database and the annotation information will also be updated to further enhance the functionalities and to provide a more reliable platform for retrieving gene expression networks in rice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figure 1.
FUNDING
The Ministry of Agriculture, Forestry and Fisheries (MAFF) of Japan; Genomics for Agricultural Innovation [RTR0002 to Y.N.]. Funding for open access charge: Genomics for Agricultural Innovation [RTR0002].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Minoru Kanehisa (Kyoto University) for providing access to KEGG pathway data. We also thank Mr Hiroya Nobori and Ms Minako Negishi (Mitsubishi Space Software Co., Ltd.) for technical support in database construction and Ms Ritsuko Motoyama (National Institute of Agrobiological Sciences) for microarray analysis.
REFERENCES
- 1.The International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 2.Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, et al. The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice Annotation Project. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice Annotation Project. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An G, Lee S, Kim SH, Kim SR. Molecular genetics using T-DNA in rice. Plant Cell Physiol. 2005;46:14–22. doi: 10.1093/pcp/pci502. [DOI] [PubMed] [Google Scholar]
- 7.Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. Generation of a flanking sequence tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 8.Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 2007;63:351–364. doi: 10.1007/s11103-006-9093-z. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, Pang J, Higashi N, Ando S, Toki S, et al. A genome-wide gain-of-function analysis of rice genes using the FOX-hunting system. Plant Mol. Biol. 2007;65:357–371. doi: 10.1007/s11103-007-9243-y. [DOI] [PubMed] [Google Scholar]
- 10.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E. ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeo K, Ishi-I J, Tamura T, Gojobori T, Tateno Y. CIBEX: center for information biology gene expression database. C. R. Biol. 2003;326:1079–1082. doi: 10.1016/j.crvi.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 15.Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat. Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61:752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- 17.Sato Y, Antonio B, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39:D1141–D1148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2012;40:D1194–D1201. doi: 10.1093/nar/gkr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao P, Jong KH, Choi D, Hwang D, Zhu J, Ronald PC. The rice oligonucleotide array database: an atlas of rice gene expression. Rice. 2012;5:17. doi: 10.1186/1939-8433-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jen CH, Manfield IW, Michalopoulos I, Pinney JW, Willats WG, Gilmartin PM, Westhead DR. The Arabidopsis co-expression tool (ACT): a WWW-based tool and database for microarray-based gene expression analysis. Plant J. 2006;46:336–348. doi: 10.1111/j.1365-313X.2006.02681.x. [DOI] [PubMed] [Google Scholar]
- 21.Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 2009;37:D987–D991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:154–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasasainagendra V, Page GP, Mehta T, Coulibaly I, Loraine AE. CressExpress: a tool for large-scale mining of expression data from Arabidopsis. Plant Physiol. 2008;147:1004–1016. doi: 10.1104/pp.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutwil M, Obro J, Willats WG, Persson S. GeneCAT–novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 2008;36:W320–W326. doi: 10.1093/nar/gkn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee TH, Kim YK, Pham TT, Song SI, Kim JK, Kang KY, An G, Jung KH, Galbraith DW, Kim M, et al. RiceArrayNet: a database for correlating gene expression from transcriptome profiling, and its application to the analysis of coexpressed genes in rice. Plant Physiol. 2009;151:16–33. doi: 10.1104/pp.109.139030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamada K, Hongo K, Suwabe K, Shimizu A, Nagayama T, Abe R, Kikuchi S, Yamamoto N, Fujii T, Yokoyama K, et al. OryzaExpress: an integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 2011;52:220–229. doi: 10.1093/pcp/pcq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obayashi T, Kinoshita K. Coexpression landscape in ATTED-II: usage of gene list and gene network for various types of pathways. J. Plant Res. 2010;123:311–319. doi: 10.1007/s10265-010-0333-6. [DOI] [PubMed] [Google Scholar]
- 28.Obayashi T, Nishida K, Kasahara K, Kinoshita K. ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 2011;52:213–219. doi: 10.1093/pcp/pcq203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato Y, Antonio BA, Namiki N, Motoyama R, Sugimoto K, Takehisa H, Minami H, Kamatsuki K, Kusaba M, Hirochika H, et al. Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol. 2011;11:10. doi: 10.1186/1471-2229-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, Minami H, Namiki N, Inukai Y, Nakazono M, et al. Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J. 2012;69:126–140. doi: 10.1111/j.1365-313X.2011.04777.x. [DOI] [PubMed] [Google Scholar]
- 31.Obayashi T, Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009;16:249–260. doi: 10.1093/dnares/dsp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38:D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39:D1114–D1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurata N, Yamazaki Y. Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 2006;140:12–17. doi: 10.1104/pp.105.063008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mihara M, Itoh T, Izawa T. SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 2010;38:D835–D842. doi: 10.1093/nar/gkp831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell. 2012;24:1848–1859. doi: 10.1105/tpc.112.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemoto T, Cho EM, Okada A, Okada K, Otomo K, Kanno Y, Toyomasu T, Mitsuhashi W, Sassa T, Minami E, et al. Stemar-13-ene synthase, a diterpene cyclase involved in the biosynthesis of the phytoalexin oryzalexin S in rice. FEBS Lett. 2004;571:182–186. doi: 10.1016/j.febslet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Cho EM, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, et al. Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J. 2004;37:1–8. doi: 10.1046/j.1365-313x.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- 45.Otomo K, Kanno Y, Motegi A, Kenmoku H, Yamane H, Mitsuhashi W, Oikawa H, Toshima H, Itoh H, Matsuoka M, et al. Diterpene cyclases responsible for the biosynthesis of phytoalexins, momilactones A, B, and oryzalexins A-F in rice. Biosci. Biotechnol. Biochem. 2004;68:2001–2006. doi: 10.1271/bbb.68.2001. [DOI] [PubMed] [Google Scholar]
- 46.Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T. Characterization of a rice gene family encoding type-A diterpene cyclases. Biosci. Biotechnol. Biochem. 2006;70:1702–1710. doi: 10.1271/bbb.60044. [DOI] [PubMed] [Google Scholar]
- 47.Shimura K, Okada A, Okada K, Jikumaru Y, Ko KW, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, et al. Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 2007;282:34013–34018. doi: 10.1074/jbc.M703344200. [DOI] [PubMed] [Google Scholar]
- 48.Okada K. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Biosci. Biotechnol. Biochem. 2011;75:1219–1225. doi: 10.1271/bbb.110228. [DOI] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 50.Xiao B, Huang Y, Tang N, Xiong L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- 51.Lenka SK, Lohia B, Kumar A, Chinnusamy V, Bansal KC. Genome-wide targeted prediction of ABA responsive genes in rice based on over-represented cis-motif in co-expressed genes. Plant Mol. Biol. 2009;69:261–271. doi: 10.1007/s11103-008-9423-4. [DOI] [PubMed] [Google Scholar]