Abstract
RNA-based next-generation sequencing (RNA-Seq) provides a tremendous amount of new information regarding gene and transcript structure, expression and regulation. This is particularly true for non-coding RNAs where whole transcriptome analyses have revealed that the much of the genome is transcribed and that many non-coding transcripts have widespread functionality. However, uniform resources for raw, cleaned and processed RNA-Seq data are sparse for most organisms and this is especially true for non-human primates (NHPs). Here, we describe a large-scale RNA-Seq data and analysis infrastructure, the NHP reference transcriptome resource (http://nhprtr.org); it presently hosts data from12 species of primates, to be expanded to 15 species/subspecies spanning great apes, old world monkeys, new world monkeys and prosimians. Data are collected for each species using pools of RNA from comparable tissues. We provide data access in advance of its deposition at NCBI, as well as browsable tracks of alignments against the human genome using the UCSC genome browser. This resource will continue to host additional RNA-Seq data, alignments and assemblies as they are generated over the coming years and provide a key resource for the annotation of NHP genomes as well as informing primate studies on evolution, reproduction, infection, immunity and pharmacology.
INTRODUCTION
Sequencing genomes has quickly become the scientific standard for being able to study any organism. The rapidly falling costs of sequencing from the development of massively parallel sequencing technologies have now made it possible for even individual laboratories to undertake whole genome efforts at unprecedented resolution and scale (1). For non-human primates (NHPs), this has resulted in genomic and transcriptomic information changing from virtually non-existent to becoming extremely expansive within the last few years (2). Complete published draft genome sequences are now available for the chimpanzee (3), gorilla (4), baboon (5) and the Indian rhesus macaque (6), along with recently completed draft genomes for the cynomolgus macaque (7) and the Chinese rhesus macaque (7). With the publication of each genome has come the increased power to make evolutionary and functional inferences. However, the annotation of these genomes has often lacked extensive evidence for the transcriptionally active units, again reflecting the historical high-cost and labor-intensive effort of cDNA sequencing, a problem affecting the annotation of both protein coding genes and the newly appreciated non-coding RNAs. The most recent estimates of the well-annotated human genome show more non-coding genes than protein coding genes (ENCODE) (8) and research has now confirmed the role of non-coding RNAs have in pre- and post-transcriptional gene regulation (9), developmental processes (10) and human disease (11). However, non-coding genes have been very limited or absent in the annotation of NHP genomes and like many protein coding genes they are inferred based on the human genome (12) rather than from species-specific evidence.
NHPs provide critical biomedical models for many aspects of human health and disease and yet the genetic basis of phenotypic traits in NHPs remains poorly understood—despite the amount of genomic data now available. Therefore, the full potential of these model organisms can only be realized with a complement of genomic information that captures both the similarities and differences to human, a requirement that is equally critical to understanding primate evolution. Most notably, comparative genomics studies strongly suggest that the significant differences between modern humans and chimpanzees are likely due at least as much to changes in gene regulation as to modifications of the genes themselves, a conjecture initially proposed by King and Wilson >30 years ago (13) and reinforced by the ENCODE results that suggest functional/regulatory roles for much of the genome that is devoid of protein coding loci.
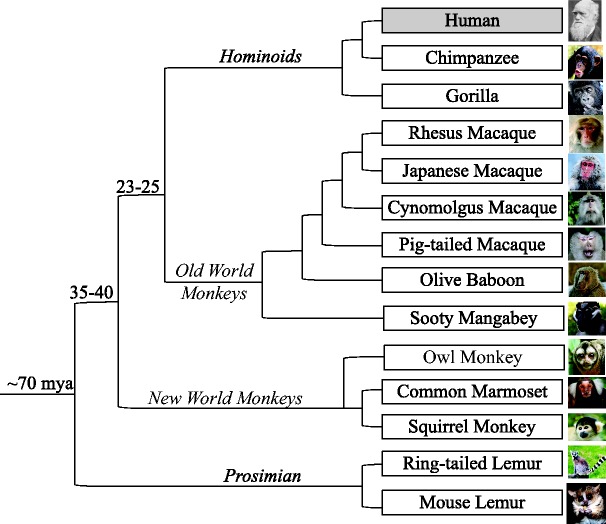
Following the 4th International Conference on Primate Genomics (Seattle, 2010), we organized a committee of investigators to assess the requirements of the research community for NHP transcriptome information; this process included representatives from many of the National Primate Research Centers, as well as experts in primate evolution from other research organizations. Based on these discussions, 13 species of NHPs were chosen for transcriptome characterization (Figure 1), with selection emphasizing their use in important biomedical models, evolutionary diversity and the status of genome sequencing. The particular importance of NHP models for studies of AIDS pathogenesis and vaccines, respiratory disease models, metabolic disorders and neurobiology led to the inclusion of multiple Macaca species, as well as geographic subspecies for the rhesus macaque and the cynomolgus macaque due to phenotypic differences noted for these regional variants. For these 15 species/subspecies, the goal for the initial sequencing effort was to capture a maximum diversity of transcripts for any one species, thereby providing a breadth of evidence for annotating transcriptionally active regions (TARs) of the respective genomes. To accomplish this, a list of 21 relevant tissues was determined that covered the range of physical and functional compartments of the animals (cf. Figure 2) and then a centrally coordinated effort was undertaken to obtain the tissues from various institutions (see ‘Materials and Methods’ section; contributing institutions are listed in ‘Acknowledgments’ section). For each species, RNA was isolated from the available tissues (with the exclusion of blood samples) and equal masses of RNA were combined to prepare the reference RNA sample that was used to generate the sequence data. (Blood RNA was not included in the general RNA composite due to the high abundance of hemoglobin RNA in such samples; therefore blood RNA will be the subject of a separate sequencing effort.) To improve functional genomics annotation for NHPs, we employed multiple methods of library preparation (14,15), thereby generating RNA-based next-generation sequencing (RNA-Seq) data characterizing coding and non-coding transcripts, delineating information on strand-specificity and enabling accurate detection of antisense transcription (Figure 2).
Figure 1.
Species of the NHPRTR. Animals were chosen to represent large evolutionary distances, encompassing hominoid, Old World and New World Monkeys and prosimians. Two geographic subspecies were included for each of the following species: rhesus macaques (Indian-origin and Chinese-origin) and cynomolgus macaques (Mauritian-origin and Indonesian-origin).
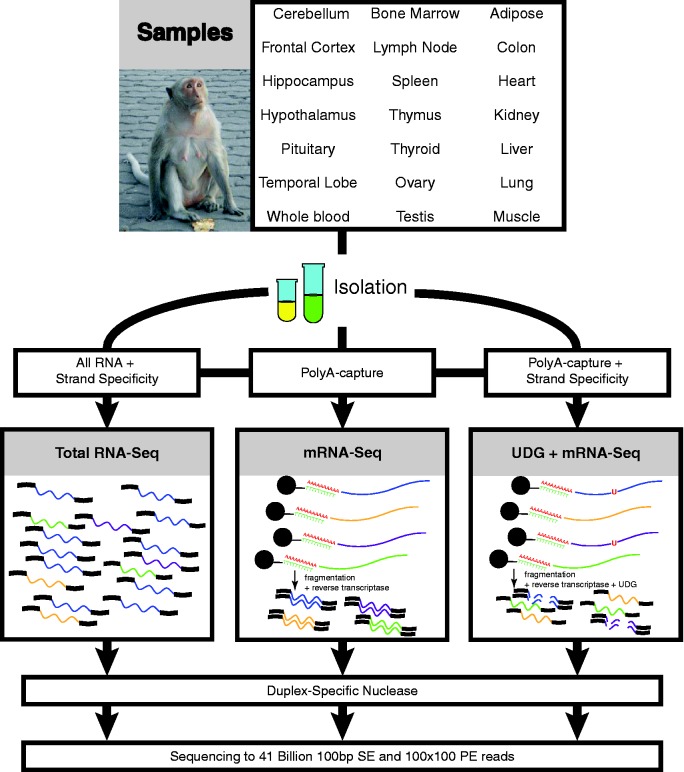
Figure 2.
Tissue sources and methods for library construction and sequencing. (Top) The tissues being sequenced (top) cover 21 regions that focus on the brain, immunological and sexual tissues as well as general tissues important for pharmacogenomics. The majority of the individual tissues will eventually be sequenced as individual libraries to examine tissue-specific expression patterns. (Middle) Three different biochemical techniques were used for preparing cDNA libraries to enable the broadest examination of the transcriptome for each species. We used an RNA-ligation method for all RNA species (Total RNA-Seq), poly-A enriched cDNA synthesis (mRNA-Seq) and another version of mRNA-Seq that maintained the Watson or Crick strand of origin for the transcript by using dUNTPs during second strand synthesis (UDG). (Bottom) All cDNA libraries were then subjected to a DNA normalization step using Duplex-specific nuclease treatment, and then all samples were clustered, sequenced, and processed using standard Illumina methods and materials, generating 41 billion reads.
We have named this effort the ‘NHP reference transcriptome resource’ (NHPRTR; online at: http://nhprtr.org), intending it to provide the community with the sequence data from the composite RNA samples and with access to derived results (processed reads, alignments, assemblies) as these become available from our own efforts as well as from others who are contributing to this central resource. Though some limited amount of NHP transcriptomic data exists (16,17), no studies or databases exist across both a large number of species and tissues, thus making the NHPRTR the most comprehensive database of primate transcriptomic information that is publicly available. Importantly, the NHPRTR is directly linked to our sample bank resource and we can provide purified RNA for the individual tissues from the species included in the resource, depending on availability.
RESULTS
Summary of primary and processed data
Our current data set contains 40.5 billion 100 nt reads from 21 tissues across 13 primate organisms (Table 1), with the majority of our data coming from 100 × 100 paired-end (PE) reads from the Illumina HiSeq2000. From the home page at http://nhprtr.org (Figure 3), our resource site is designed to provide easy access to many resources, including pages that describe the overall goals of the project, its current status, contact information, external links and also the link to the data page. The data page hosts all of the raw data from the sequencing of the various species and each of their library preparations, with a file name that represents the provenances of the data generation. For example, the PE reads sequences from the Baboon UDG library called ‘HCT20960’ sequenced on lane five, appear as BAB_UDG_HCT20960_L005_R1.fastq.gz and BAB_UDG_HCT20960_L005_R2.fastq.gz. Finally, under each set of data, we have posted md5sums of each of the files, so users can readily confirm their accurate receipt of the data after download.
Table 1.
Summary of current data in NHPRTR. The 40.5 billion reads span three different library preparation methods and two sequencing instruments (GAIIx and the HiSeq2000)
| Species | File size (GB) | HiSeq2000 (2 × 100 nt paired-end reads) GAII (100 nt single-end reads) |
|||
|---|---|---|---|---|---|
| Protocol | Number of read pairs | Protocol | Number of reads | ||
| Baboon | 973 | mRNA-seq | 955 573 799 | mRNA-seq | 71 477 607 |
| UDG mRNA-seq | 918 735 897 | UDG mRNA-seq | 67 763 503 | ||
| Total RNA-seq | 151 524 634 | ||||
| Chimpanzee | 94.9 | Total RNA-seq | 198 954 000 | ||
| 399.1 | UDG mRNA-seq | 836 864 082 | |||
| Cynomolgus Macaque Indochinese | 948 | mRNA-seq | 923 307 160 | mRNA-seq | 72 016 960 |
| UDG mRNA-seq | 894 367 594 | UDG mRNA-seq | 63 820 198 | ||
| Total RNA-seq | 157 762 299 | ||||
| Cynomolgus Macaque Mauritian | 656 | mRNA-seq | 503 249 742 | mRNA-seq | 90 166 271 |
| UDG mRNA-seq | 557 450 722 | UDG mRNA-seq | 91 262 189 | ||
| Total RNA-seq | 176 108 975 | ||||
| Gorilla | 98.5 | Total RNA-seq | 206 526 535 | ||
| 422.6 | UDG mRNA-seq | 886 261 413 | |||
| Japanese Macaque | 986 | mRNA-seq | 942 269 530 | mRNA-seq | 77 740 433 |
| UDG mRNA-seq | 943 158 996 | UDG mRNA-seq | 72 925 864 | ||
| Total RNA-seq | 181 184 542 | ||||
| Marmoset | 128.8 | Total RNA-seq | 269 969 905 | ||
| 418.9 | UDG mRNA-seq | 878 369 246 | |||
| Mouse Lemur | 97.5 | Total RNA-seq | 204 494 231 | ||
| 378.9 | UDG mRNA-seq | 794 659 816 | |||
| Pig-tailed Macaque | 951 | mRNA-seq | 867 009 248 | mRNA-seq | 54 292 043 |
| UDG mRNA-seq | 991 993 458 | UDG mRNA-seq | 54 668 320 | ||
| Total RNA-seq | 131 548 564 | ||||
| Rhesus Macaque Chinese | 700 | mRNA-seq | 644 468 744 | mRNA-seq | 77 142 089 |
| UDG mRNA-seq | 661 177 666 | UDG mRNA-seq | 75 142 089 | ||
| Total RNA-seq | 121 570 595 | ||||
| Rhesus Macaque Indian | 1331.2 | mRNA-seq | 1 716 083 364 | mRNA-seq | 84 892 037 |
| UDG mRNA-seq | 704 493 397 | UDG mRNA-seq | 70 346 332 | ||
| Total RNA-seq | 168 710 995 | ||||
| Ring-tailed Lemur | 104.8 | Total RNA-seq | 219 647 886 | ||
| 398.7 | UDG mRNA-seq | 835 972 568 | |||
| Sooty Mangabey | 106 | Total RNA-seq | 222 192 769 | ||
| 424.4 | UDG mRNA-seq | 889 864 522 | |||
| Total | 9618.3 | 18 667 116 290 | 2 112 066 539 | ||
| Total number of reads | 39 446 299 119 | ||||
Although the sequencing for all species is not identical, due to increased use of the higher output HiSeq2000 in later species, we represent both total RNA and polyA-enriched RNA preparations for all species.
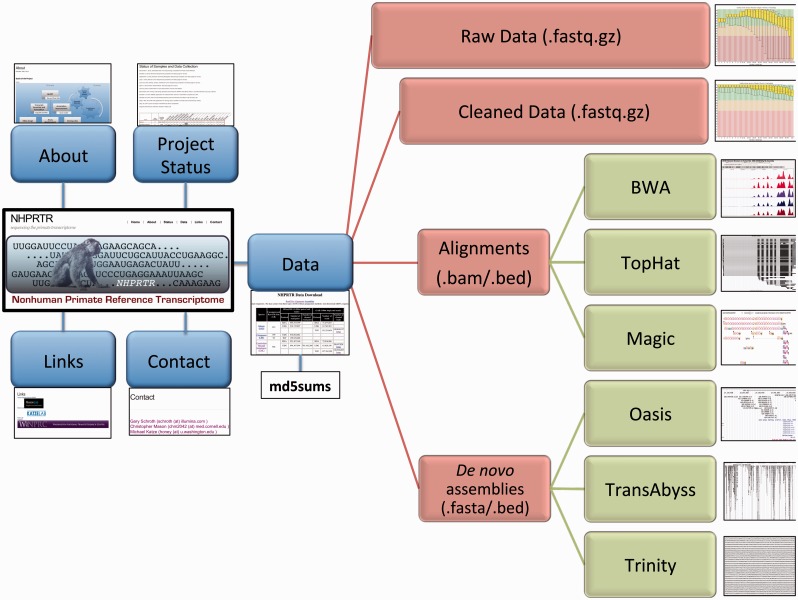
Figure 3.
Organization of the NHPRTR. We have designed our database and the interface page to give users a clear sense of the goals, organization and data types present. Pages include the project background (about.html), the latest updates (status.html), connections to other sites (links.html), contact information (contact.html) and the data from the NHPRTR (data.html, including md5sums). From the data page (middle), users can access the raw data or various forms of the processed data, including cleaned data, alignments (using BWA, TopHat or Magic) and assemblies (Oases, TransAbyss and Trinity). The data page will continually update as data are submitted and as work is completed.
Our primary data analysis and quality checking have shown that our data are of very high quality (Supplementary Figure S1), with a median Quality Score (Q-score) consistently >34 (>99.95% accuracy) across the length of the reads. Also, we used the tool Stitch (18) to check the overlap of the reads and found that the insert size of the cDNA libraries were within the expected range of 140–160 bp, since the mode of the distribution of the overlap of the PE (100 × 100) libraries was near 40–60 (Supplementary Figure S2). Finally, the data distribution page also provides the output files from the FASTQC toolkit, to allow a deeper examination of the read statistics and qualities (19).
Once a species is sequenced and quality checked, the NHPRTR site also hosts a second version of the primary data. This second set is a ‘cleaned version’ which is generated for use in algorithms that are especially sensitive to sequence errors, such as de novo transcript assemblers and genome assemblers (Supplementary Figure S3). We first trim all reads for low quality (<Q20), remove any remaining adapter sequences and any lengthy polyA/T stretches (>6 homopolymers) using Flexbar, in order to eliminate bad quality reads and the sequences from the ends of polyA tails or low complexity regions. We then align all reads to the known primate sequences for mtDNA and rRNA and exported these to a separate alignment files. We found that these steps remove between 3% and 10% of the data. These files can save significant time for researchers who want to begin with even higher quality data and who do not wish to focus on the mitochondrial or ribosomal sequences.
Alignments and data visualization
As the gene models for each species improve, it is often useful to gauge the state of these emerging data in relation to the best defined gene annotation set available—the human genome. To enable such work, the NHPRTR hosts an alignment to the human genome (hg19 and hg18) using Burrows–Wheeler Aligner (BWA) (20) and can all be readily viewed within the UCSC genome browser from a direct link on http://nhprtr.org. While we recognize that using the human genome as an alignment reference for distant phylogenetic species is not ideal, we still provide these alignments for several reasons. First, the human genome is the best annotated genome across primates and it hosts a wealth of other regulatory and functional data linked to the genomic coordinates. Second, the data can already be useful as a comparison of gene structures in expressed areas, placing genes in syntenic blocks and helping to define orthologous gene sets. Third, some species in the NHPRTR database have no genome yet sequenced. Finally, even though sequence divergence will decrease mapping rates, the alignments still provide a basic orthologous expression map across all species.
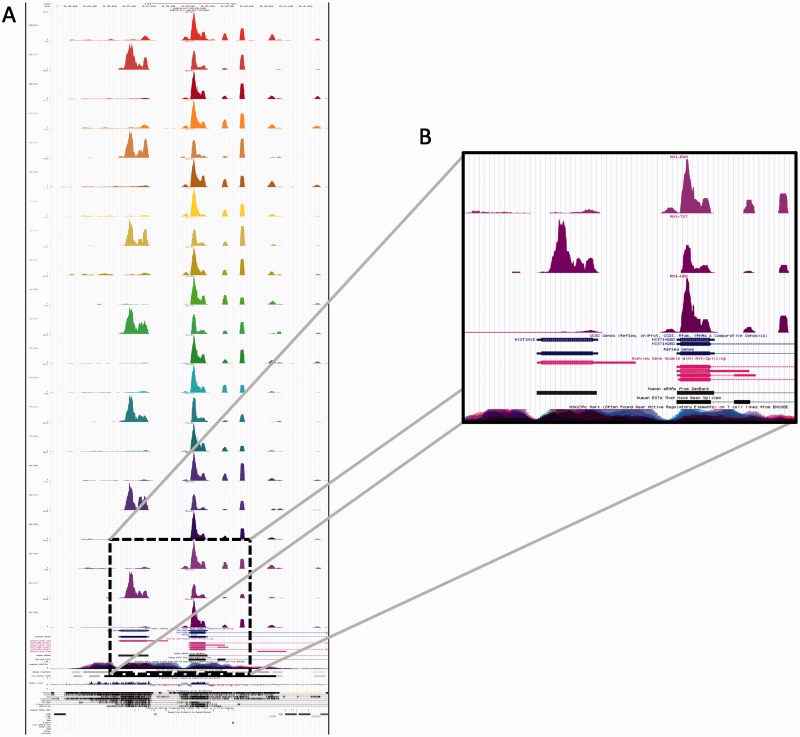
We observed that these human alignment data generated several immediate results. First, users can browse to any given human gene of interest and gauge the gene structure and rough expression level of that gene. Second, any hypothesized changes in gene structure, such as shortening, lengthening or splicing changes, can be visualized and compared to human structures. Third, the differences in the types of RNA-Seq can readily show the benefit of using multiple biochemical methods for the examination of a transcriptome (Figure 4). For instance, the detection of non-poly-adenylated transcipts such as snoRNAs or some histones can be readily seen in the Total RNA prep, whereas they are missing from the two mRNA preps.
Figure 4.
Browsable tracks. (A) We used BWA aligner to create cross-species maps of expression, based on the alignment to orthologous sequences of the human genome. Here we show the three library preparation methods (TOT, UDG, RNA), with one in each track for seven species. (B) The insert shows how the Total RNA preparation method (middle expression track) can more readily discern non-poly-adenylated genes, such as the histone genes.
Ongoing database work and analysis plans
As described here, this large-scale, EST-like resource of 13 species/subspecies of NHPs across 21 tissues is immensely useful for primate researchers, evolutionary biologists, immunologists and neurobiologists. With the addition of the Squirrel Monkey, Owl Monkey and other tissue-specific sequencing, we anticipate having ∼100 billion reads from 15 primates when sequencing is completed in 2013. We plan to sequence individual tissues from the Indian-origin rhesus macaque from animals at different stages following SIV infection and also perform tissue-specific sequencing using different cDNA methods. Taken together, these data will create an unprecedented depth of expression and single-base resolution expression data for all of these species’ tissues. Most significantly, the different types of biochemistries utilized for cDNA synthesis and RNA preparation for sequencing will create a broad, comprehensive profile (polyA and total RNA) of the transcriptome for each tissue and each species.
Several ongoing analysis efforts from these data will be posted to the NHPRTR site, leveraging a variety of aligners and assemblers. First, as relevant published work in NHP transcriptomes appear (21), we will link to them on our site. Next, additional alignments from the AceView aligner (22) will be added, which will be very useful for defining the non-coding RNAs of the transcriptome and also the exon–intron junctions, along with the outputs from Tophat and BWA. We will also host the results of several de novo transcriptome assemblies as they are completed, based on the different libraries and available PE and single-end reads as described earlier. At time of submission, we are hosting preliminary de novo assembly results for the Mauritian cynomolgus macaque from pilot studies using Oases (23), TransAbyss (24) and Trinity (25), performed with subsets of the data (cf. Data under http://nhprtr.org). These assemblies are also linked to the reference genome for each species for species-specific browsing. Notably, our early efforts revealed that the construction of the de novo transcriptome assemblies can be a very memory-intensive process (Supplementary Figure S3), which often required hundreds of gigabytes RAM. Thus, in an effort to help researchers utilize these data in large-scale computing environments, we are also hosting these data on the Blacklight 32TB memory node (blacklight.psc.teragrid.org) on the Extreme Science and Engineering Discovery Environment (XSEDE). We anticipate that having various means of accessing the primary, processed and assembled reads in multiple environments will ensure the broadest utilization of these data.
In summary, we have designed the NHPRTR site to utilize familiar tools and formats from the genomics community and the combination of several library preparations and bioinformatic tools in the same resource have already created thousands of requests to examine and download these data (Supplementary Figure S4). We encourage the use of the data by the community and will assist investigators in hosting their results if they wish to contribute to the resource and in reciprocity, we also have a section with links to data from other published primate RNA-Seq studies. Moreover, a main goal in generating these data was to provide a rich resource for species-specific alignment, thereby generating improved gene models for NHP genomes and this is realized by own ongoing efforts as well the gene annotation and prediction pipelines at ENSEMBL (B. Aken, personal communication). These data will also be helpful in answering a variety of questions pertaining to the complexity of transcriptome, including: new TARs, conservation/evolution of specific splicing sites, RNA editing events, UTR structures, and gene boundaries and content. In summary, the NHPRTR represents an immensely useful and timely addition to the genome sequences of these important species, a key hub for these species’ RNAs and their matching transcriptomic data and an invaluable resource for genomes that will eventually be sequenced.
MATERIALS AND METHODS
Tissue samples
Source tissues for the resource were generally obtained from animals that were being euthanized either for compassionate reasons due to failing health or as part of an existing research protocol; all veterinary procedures were approved under the local Institutional Animal Care and Use Committee (IACUC). Tissue specimens were preserved in RNAlater® (Life Technologies) at the time of collection and frozen at −80°C. Tissues for gorilla, mouse lemur and ringtail lemur derived from frozen specimens previously collected at the time the individual animals were euthanized; these tissues were either transferred into RNAlater or homogenized in TRIzol® Reagent (Life Technologies) and frozen at −80°C. All frozen samples were shipped to the University of Washington and the RNA isolated under a standard protocol using TRIzol extraction and purification with RNeasy® columns (QIAGEN). Isolated RNA was characterized by absorbance spectroscopy to ensure the absence of contamination by protein or phenol and then analyzed by capillary electrophoresis to furnish an RNA Integrity Number (RIN) using an Agilent Bioanalyzer®. RNA concentrations for individual tissue RNA samples were based on integrated fluorescence intensity in the Bioanalyzer runs, calibrated against an RNA standard. For a species or subspecies, the reference sample combined equal masses of RNA from all the tissues. The number of available tissues varied among the species; whenever possible tissues were used from a single female individual and only was obtained from a second individual (see Supplementary Table S1). The final composition of each reference sample as well as the RIN value for the individual tissue RNA components is available at the resource website (http://nhprtr.org).
Library preparations
Three different types of sequencing libraries were prepared from the reference samples. These were as follows: (i) non-directional mRNA-Seq, (ii) directional mRNA-Seq based on dUTP strand-marking and (iii) directional Total RNA-Seq, based on RNA-ligation to the initial RNA fragments which preserves their strandedness. In all the cases, the initial cDNA library was ‘normalized’ using a Duplex-Specific Nuclease Protocol (DSN) which removes high-abundance transcripts such as ribosomal molecules that would otherwise dominate the reads from the Total RNA-Seq libraries. The majority of sequencing for all species was done on the Illumina HiSeq2000 at Illumina or Weill Cornell Medical College (WCMC), with additional GAIIx sequencing performed at Illumina.
Standard mRNA-Seq protocol
The standard mRNA-Seq library preparations were done using established Illumina methods for mRNA-Seq (Part #RS-100-0801). Briefly, poly A+ RNA is purified from 100 ng of total RNA with oligo-dT beads. Purified mRNA is then fragmented with divalent cations at elevated temperature. First strand cDNA synthesis is performed with random hexamer priming and reverse transcriptase. Second strand cDNA synthesis is performed using RNAseH and DNA PolI. Following cDNA synthesis, the double stranded products are end repaired, followed by addition of a single ‘A’ base and then ligation of the Illumina PE adaptors. For this study, the ligation products were purified using gel electrophoresis. The target size range for these libraries was ∼250 bp on the gel such that the final library for sequencing would have cDNA inserts with sizes of ∼150 bp long. Following gel purification the adapter ligated cDNA is then amplified with 15 cycles of PCR. This initial library was then subject to DSN normalization and additional rounds of PCR as described below for the Total RNA-Seq protocol.
Directional (UDG) mRNA-Seq protocol
The directional mRNA-Seq library preparations were done using the variant offered by Illumina (Part # RS-122-2303). Briefly, poly A+ RNA is purified from 100 ng of total RNA with oligo-dT beads. Purified mRNA is then fragmented with divalent cations under elevated temperature. First strand cDNA synthesis is performed with random hexamer primers and reverse transcriptase. Second strand cDNA synthesis is performed using RNAseH, dATP, dCTP, dGTP, dUTP and DNA PolI. Following cDNA synthesis, the products are end repaired, a single ‘A’ base is added and then the Illumina PE adaptors are ligated on to the cDNA products. The libraries are then amplified with 15 cycles of PCR as before, except in this case the strands that contain dUMP do not amplify and thus the products of the PCR process retain the original strand information. For this study, the ligation products were purified using gel electrophoresis. The target size range for these libraries was ∼300 bp on the gel such that the final library for sequencing would have cDNA inserts with sizes of ∼200 bp long. This initial library was then subject to DSN normalization and additional rounds of PCR as described below for the Total RNA-Seq protocol.
RNA-ligation-based directional total RNA-Seq protocol with DSN
The directional Total RNA library is constructed with a modified version of the Illumina directional mRNA-Seq sample preparation protocol, however no poly-A selection is used in a Total RNA-Seq prep. Briefly, 100 ng of total RNA is fragmented with divalent cations under elevated temperature. The ends of the fragmented RNA are treated with phosphatase to remove all 5′- and 3′-phosphate groups, followed by modification with polynucleotide kinase. This process insures that every RNA molecule contains a 5′-mono-phosphate group and a 3′-hydroxyl group. A pre-adenylated oligo is then ligated to the 3′-end of these RNA fragments, followed by the ligation of an RNA oligo to the 5′-end of the RNA. Following ligation of these adapter oligos, the RNA is reverse transcribed and amplified with 15 cycles of PCR to create the initial RNA-Seq library. Ribosomal RNA depletion from the initial RNA-Seq library is carried out following Illumina’s published protocol. Briefly, 100 ng of amplified PCR products are denatured at 94°C for 5 min in 1× hybridization buffer (50 mM HEPES, 0.5 M NaCl) followed by incubation at 68°C for 5 h; then 2 U of the DSN Enzyme (available from Evrogen) is added at 68°C for 25 min to digest double stranded DNA. Following DSN digestion the remaining undigested, single-stranded molecules are enriched with 15 more cycles of PCR.
Alignment methods
We used several alignments strategies on the data, with an initial focus on the alignment of the various species on the human genome. For an extremely conservative view of cross-species mapping, we used the BWA (20) and removed any sub-optimal matches (X0==1) and also removed any reads that were one edit distance away from mapping somewhere else in the genome (X1==0) field. These parameters reduced issues with paralogs and segmental duplications. For a broader alignment method, we used the AceView Magic aligner (22) and Tophat (26) (default settings) to generate mapping rates for each library of each species. The Magic AceView aligner uses a compressed data format for rapid processing and then uses a seed-and-extend algorithm based on sequence complexity, boundary detection for splicing, a scoring matrix for alignment and a mapping hierarchy to assign the reads to the most likely location in the genome. Specific bash commands and shell scripts that were used in the analysis are posted online at nhprtr.org and also in Supplementary Data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–4 and Supplementary Methods.
FUNDING
National Institutes of Health Office of Research Infrastructure Programs [R24RR032341]; Washington National Primate Research Center [P51RR000166 to Katze Laboratory]; [1R01NS076465-02 to Mason Laboratory]; XSEDE super-computing cluster [MCB120116]; STRIDE Center for Systems and Translational Research for Infectious Diseases at the University of Washington; National Center for Biotechnology Information (NCBI) [to J.T.-M. and D.T.-M.]; Intramural Research Program of the NIH, National Library of Medicine [partial]. Funding for open access charge: NIH and NCRR grants.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Illumina, Inc. for contributing almost all of the resources and reagents needed for completing the sample preps, sequencing and primary data analysis. Many people at Illumina helped create the libraries and sequencing data but we would especially like to recognize the efforts of Shujun Luo, Irina Khrebtukova, David Silva, Cindy Chen, Robin Li and Hang Pham. Tissues were obtained as research resources from the following centers: Washington National Primate Research Center, Wisconsin National Primate Research Center, Oregon National Primate Research Center, Yerkes National Primate Research Center, Southwest National Primate Research Center, the Duke University and the Duke Lemur Center; the Keeling Center for Comparative Medicine and Research, the North Carolina Zoo and Covance Inc. The Weill Cornell Medical College Epigenomics Core Facility provided support for use of their sequencing machines and technical assistance during sequencing. Finally, we would like to thank Bronwen Aken, Paul Flicek and Steve Searle from ENSEMBL for coordination of processed data also on their site.
REFERENCES
- 1.Perry GH, Reeves D, Melsted P, Ratan A, Miller W, Michelini K, Louis EE, Jr, Pritchard JK, Mason CE, Gilad Y. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol. Evol. 2012;4:126–135. doi: 10.1093/gbe/evr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 4.Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babbitt CC, Tung J, Wray GA, Alberts SC. Changes in gene expression associated with reproductive maturation in wild female baboons. Genome Biol. Evol. 2012;4:102–109. doi: 10.1093/gbe/evr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhesus Macaque Genome Sequencing and Analysis Consortium et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 7.Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, et al. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat. Biotechnol. 2011;29:1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 8.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van IJcken W, Grootegoed JA, Gribnau J. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- 10.Ahfeldt T, Schinzel RT, Lee YK, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat. Cell Biol. 2012;14:209–219. doi: 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 14.Luo S, Smith GP, Khrebtukova I, Schroth GP. Total RNA-seq: complete analysis of the transcriptome using Illumina sequencing-by-synthesis sequencing. In: Harbers M, Kahl G, editors. Tag-Based Next Generation Sequencing. 1st edn. Wiley-VCH Verlag GmbH & Co. KGaA; 2012. pp. 367–381. [Google Scholar]
- 15.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry GH, Melsted P, Marioni JC, Wang Y, Bainer R, Pickrell JK, Michelini K, Zehr S, Yoder AD, Stephens M, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 18.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Alcántara A, Ballesteros E, Feng C, Rojas M, Koshinsky H, Fofanov VY, Havlak P, Fofanov Y. PIQA: pipeline for Illumina G1 genome analyzer data quality assessment. Bioinformatics. 2009;25:2438–2439. doi: 10.1093/bioinformatics/btp429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babitt CC, Fedrigo O, Pfefferle AD, Boyle AP, Horvath JE, Furey TS, Wray GA. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol. Evol. 2010;2010:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1):S12.1–S14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. De novo assembly and analysis of RNA-seq data. Nat. Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 25.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]