Abstract
Whipple's disease is considered a rare chronic disease with a broad spectrum of clinical manifestations. Several antibiotics have been used for the treatment of this disease, and the current reference treatment was determined empirically on the basis of only a few clinical observations. Patients should be treated for months, and many relapse after antibiotic withdrawal. We report here the first extensive study on the susceptibilities of three reference strains of Tropheryma whipplei to antibiotic in cell culture by using a real-time PCR assay as previously described. We found that doxycycline, macrolides, ketolides, aminoglygosides, penicillin, rifampin, teicoplanin, chloramphenicol, and trimethoprim-sulfamethoxazole were active, with MICs ranging from 0.25 to 2 μg/ml. Vancomycin was somewhat active at an MIC of 10 μg/ml. We found heterogeneity in the susceptibility to imipenem, with one strain being susceptible and the two other strains being resistant. Cephalosporins, colimycine, aztreonam, and fluoroquinolones were not active. We also demonstrated that a combination of doxycycline and hydroxychloroquine was bactericidal. This combination has been shown to be active in the treatment of patients suffering from chronic infections with Coxiella burnetii, a bacterium that is also found intracellularly in acidic vacuoles. We believe, then, that this combination therapy should be further evaluated in clinical trials for the treatment of Whipple's disease.
Whipple's disease is a chronic systemic infection affecting mostly middle-aged males (28, 33). Although first described in 1907 (35), it is a rare disease with fewer than 1,000 cases reported (8). There is a wide heterogeneity of clinical manifestations, including a prodromal period of polyarthritis, fatigue, weight loss, and anemia, which is followed by a progressive syndrome of abdominal pain and distention, steatorrhea, and severe cachexia (15). Other clinical manifestations that have been reported include endocarditis, myocarditis, pericarditis, and neurological signs.
The agent of Whipple's disease, Tropheryma whipplei, has recently been isolated in a cell culture from a patient with endocarditis (24). This has enabled a more complete characterization of the organism which is an aerobic, rod-shaped, gram-positive, filamentous bacterium (0.5 to 2 μm) that can be found both intracellularly and extracellularly (8). Of note is the fact that these bacteria grow slowly in acidic vacuoles of cells (12), a finding which has also been described with other strict intracellular bacteria—in particular, Coxiella burnetii—that also leads to chronic infections that are difficult to treat (19). Moreover, during the cell culture the bacteria leads to the production of cords in extracellular medium (4, 8, 15, 23). It has therefore been suggested that the bacteria could have an extracellular life cycle (29).
Phylogenetically, T. whipplei is classified as a member of the Actinomycetes and has been placed between the genus Cellulomonas and the Actinomycetes clade (16). The complete genome of the bacteria has recently been sequenced, which has confirmed its phylogenetic position (1, 27).
Definitive diagnosis of Whipple's disease is now possible by various methods, including immunohistochemistry and PCR assays for various target genes on biopsy samples (8). The PCR assay has become an important diagnostic tool for the diagnosis of Whipple's disease, especially in patients with unusual presentations and in patients in which the diagnosis cannot be confirmed histologically.
The currently recommended treatment for Whipple's disease has been determined empirically. The reference treatment is a combination of streptomycin (1 g) and benzylpenicillin (penicillin G; 1.2 million units) for 14 days and thereafter oral cotrimoxazole (trimethoprim-sulfamethoxazole; 160 mg/800 mg twice daily) for 1 year (14, 31). With this treatment regimen, however, relapses have been reported after cessation of antibiotic therapy (5, 11, 17). Dykman et al. have suggested that this may be because trimethoprim-sulfamethoxazole is only bacteriostatic despite the high intracellular concentrations the drug achieves (5). The use of bactericidal antibiotics may then be more appropriate in the treatment of Whipple's disease (5, 8).
We recently developed a new genomic assay for the determination of antibiotic susceptibilities by using the LightCycler instrument (18) and found that T. whipplei was naturally resistant to fluoroquinolones due to specific mutations within the DNA gyrase gene (18). In the experiments we now report, we used this technique to evaluate the activity of other antibiotics against three isolates of T. whipplei. Previous studies have shown that alkalinization of the acidic vacuoles in which C. burnetii and Staphylococcus aureus are found promotes bactericidal activity in vitro (19, 25). Since T. whipplei also occurs in acidic vacuoles, we evaluated the bactericidal effect of a combination of doxycycline and alkalinizing agents on the organism. Such a combination has previously been shown to be active against C. burnetii and S. aureus (19, 25).
MATERIALS AND METHODS
T. whipplei strains
Three T. whipplei isolates obtained in our laboratory and cultured in MRC5 fibroblast cells (ATCC number CCL-171) were tested in our study: Twist strain, isolated from a heart valve (24) and passaged 50 times; Endo-5 (6), isolated from a heart valve and passaged 44 times; and Slow2 (6), isolated from a duodenal biopsy and passaged 33 times. These data are presented in Table 1.
TABLE 1.
T. whipplei strains and patient characteristics
| Patient sex (age [yr]) | Strain | Symptoms | Biopsy sample | Antibiotic treatment before sampling | No. of passages |
|---|---|---|---|---|---|
| Male (42) | Twist | Endocarditis | Cardiac valve | Gentamicin plus ampicillin (4 wk), ciprofloxacin (several wk) | 50 |
| Female (74) | Slow2 | Diarrhea, arthralgia | Duodenum | None | 33 |
| Male (63) | Endo-5 | Endocarditis | Blood | Gentamicin plus amoxicillin (3 wk) | 44 |
T. whipplei-infected MRC5 cells were grown in 150-cm2 culture flasks (Falcon; Beckman) incubated at 37°C in a 5% CO2-enriched atmosphere. Minimum essential medium (Life Technologies/Gibco-BRL and Cergy Pontoise) supplemented with 10% fetal bovine serum and 2 mM l-glutamine (Gibco) was used as the incubation medium (15). Cell infection rates were monitored three times a week by examining cells scraped from the flasks and stained with Gimenez stain (13).
Antibiotic solutions
The antibiotics used in the study were tested in serial twofold dilutions as follows: doxycycline (0.5 to 8 μg/ml; Pfizer, Neuilly, France), levofloxacin (2 to 8 μg/ml; Hoechst Marion Roussel, Romainville, France), ofloxacin (0.5 to 8 μg/ml; Diamant, Puteaux, France), ciprofloxacin (0.5 to 8 μg/ml; Bayer Pharma, Sebs, France), erythromycin (0.06 to 10 μg/ml; Abbot, Rungis, France), telithromycin (0.5 to 8 μg/ml; Hoechst Marion Roussel), thiamphenicol (0.06 to 32 μg/ml; Sanofi Winthrop, Gentilly, France), rifampin (0.5 to 8 μg/ml; Cassenne, Puteaux, France), trimethoprim-sulfamethoxazole (0.06 to 32 μg/ml; Roche, Paris, France), gentamicin (0.25 to 10 μg/ml; Dakota Pharm, Creteil, France), amoxicillin (0.25 to 8 μg/ml; SmithKline Beecham, Nanterr, France), streptomycin (0.25 to 8 μg/ml; Diamant), ceftriaxone (0.25 to 100 μg/ml; Roche), penicillin G (0.06 to 10 μg/ml; Diamant), vancomycin (1 to 100 μg/ml; Dakota Pharm), clarithromycin (1 to 2 μg/ml; SmithKline Beecham), teicoplanin (Marion Merrell Dow, Paris, France), imipenem (0.25 to 10 μg/ml; Dakota Pharm), aztreonam (1 to 100 μg/ml; Sanofi Winthrop), cephalotin (1 to 100 μg/ml; Panapharma, Luitré-Fougeres, France), chloramphenicol (1 to 2 μg/ml; Coger, Paris, France), colimycin (Roche), hydroxychloroquine (1 to 2 μg/ml; Sanofi Synthelabo, Paris, France), and NH4Cl (1 to 2 μg/ml; Coger, Paris, France).
Stock solutions were prepared according to the manufacturers' instructions and stored at −80°C until used. Working solutions were prepared extemporaneously by diluting stock solutions in minimal essential medium supplemented with 2 mM glutamine and 5% fetal bovine serum.
Antibiotic susceptibility testing of T. whipplei isolates
We previously described a real-time PCR assay used to determine antibiotic susceptibility (18). Briefly, when cells were heavily infected, usually after 3 weeks of incubation, the cell supernatant was discarded from the flask, and the infected MRC5 cells were detached by using sterile glass beads with 5 ml of fresh medium. Cells were lysed by sonication (three 30-s sonications in ice at 60 mV) and centrifuged (15 min at 20,000 × g) to discard cell debris, and the supernatant was diluted 1:1,000 in culture medium. This suspension was used to infect confluent MRC5 monolayers in 48-well microtiter plates (D. Dutcher, Brumath, France), which were incubated at 37°C in 5% CO2. Infected cells in three different wells were harvested every 3 days from days 0 to 12 postinfection. The growth kinetics of T. whipplei in MRC5 cells was determined by determining the numbers of genome copies in each cell suspension by quantitative PCR.
For antibiotic susceptibility testing, dilutions of antibiotics (prepared as described above) were added at serial twofold dilutions to the culture media of individual tissue culture plate wells after 48 h of incubation, in duplicate. Antibiotic-free wells served as growth controls, whereas uninfected MRC5 cell wells served as negative controls. During the antibiotic challenge experiment, cell cultures from wells were harvested every 3 days over a 12-day period, and cell suspensions were frozen at −80°C until DNA was extracted for quantitative PCR assays. The lack of toxicity of antibiotics to MRC5 cells was determined by examination of cell monolayers under an inverted microscope (Zeiss Axiovert 25; Carl Zeiss) at the time cell cultures were harvested. The MIC was defined as the lowest antibiotic concentration at which complete inhibition of bacterial growth was determined by measurement of DNA copies by quantitative PCR assay. The number of DNA copies present after 12 days was compared to the number of DNA copies at day 0 of the experiment. Experiments were carried out on three separate occasions, each time in duplicate, to determine consistency of results.
Controls used for antibiotic activity
Escherichia coli ATCC 8739 and S. aureus CIP ATCC 49976 were obtained from the Pasteur Institute (Institut Pasteur, Marnes La Coquette, France) and used as antibiotic test controls (18). Antibiotic activities were determined by using Mueller-Hinton agar (bioMérieux) incubated at 37°C for 18 h. The activities of the various dilutions of the antibiotics described above were determined after 15 days of incubation at 37°C.
Bactericidal activity of antibiotics
The bactericidal activity or postantibiotic effect of doxycycline in combination with hydroxychloroquine was determined by subculturing 100 μl of harvested cell suspensions, collected on day 12 of the experiment, onto fresh cell cultures containing antibiotic-free culture medium. After 7 and 10 days of culture, sample cells were harvested as described above for real-time PCR and Gimenez staining. The bactericidal effect or the postantibiotic effect was defined as the lack of regrowth of bacteria after 10 days of subculture.
Quantitative PCR assay by using a LightCycler
T. whipplei DNA was extracted from 100-μl aliquots of infected MRC5 cells by using the MagNA Pure LC DNA Isolation Kit III (Roche Applied Science, Penzberg, Germany) as described by the manufacturer. The extracted nucleic acid was resuspended in a final volume of 100 μl and stored at −20°C until used in the quantitative PCR assay.
PCR was performed by using a LightCycler instrument (Roche Biochemicals, Mannheim, Germany) with 20-μl volume glass capillaries. The PCR mixture (20 μl) contained 2 μl of extracted DNA, 13.2 μl of H2O, 1.6 μl of MgCl2 (25 mmol), 1 μl (i.e., 10 pmol) of the forward primer ITS (5′-CCGAGGCTTATCGCAGATTG-3′), 1 μl (i.e., 10 pmol) of the reverse primer ITS (5′-GGTGACTTAACCTTTTTGGAG-3′) (7), and 2 μl of DNA-Master Hybridization Probes (Roche Diagnostics) containing Taq DNA polymerase, reaction buffer, a deoxynucleoside triphosphate mixture, and 3 mmol of MgCl2 (10-fold concentrated). The PCR was performed with an initial denaturation at 95°C for 8 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 5 s, and extension at 72°C. A calibration curve for DNA quantification was established by amplifying 10-fold serial dilutions of DNA extracted from the primary T. whipplei inoculum used to infect MRC5 cells.
The specificity of the PCR products was verified by sequencing as previously described (18).
RESULTS
Sensitivity and specificity
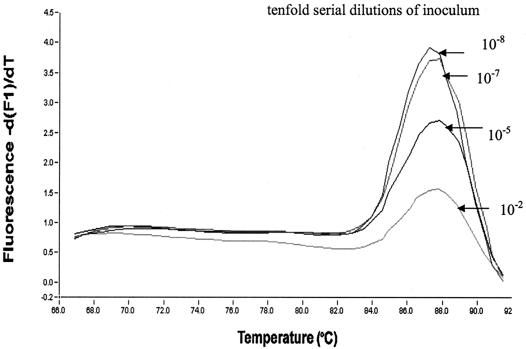
Figure 1 shows the melting curves obtained with standard concentrations of a previously quantified inoculum of T. whipplei (108) and the specificity of the PCR product (a single sharp melting temperature obtained at ca. 87.5°C), as determined by using a standard calibration curve obtained with 10-fold serial dilutions of T. whipplei. The standard curve was determined in each experiment to enable the results of all experiments to be correlated. After resolution by agar electrophoresis, all T. whipplei PCR products showed a single band fragment. Sequences of this unique band were always 100% identical to the sequence of the ITS gene of T. whipplei (GenBank accession number AF248312).
FIG. 1.
Melting curves obtained with standard concentrations of T. whipplei by PCR by measuring the amount of fluorescence (Df/dT) with a LightCycler. The specificities of the PCR products are shown by detection of a single peak at 88°C.
Growth kinetics of T. whipplei as determined by using real-time PCR
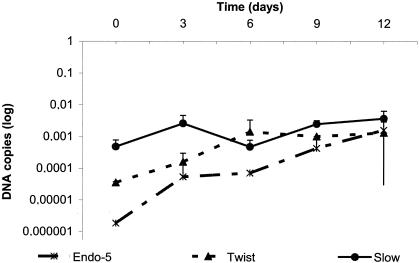
Fibroblast cells present a contact inhibition when monolayers are confluent, and thus the numbers of cells in each well were approximately the same (Fig. 2). The viability of cells was verified by dye staining. The growth of T. whipplei isolates was slow, with 2.3 ± 3.8 log increases in bacterial concentration over 12 days of incubation for Twist strain, 4.3 ± 4.65 log for Endo2 strain, and 1.8 ± 5.4 log for Slow2 strain. During this period, the doubling time of T. whipplei Twist was determined to be 35 h, whereas that of Endo2 was 32 h and that of Slow2 was of 48 h. Thereafter, bacterial growth plateaued.
FIG. 2.
Growth kinetics of T. whipplei strains (Twist, Endo-5, and Slow2) cultured in MRC5 cells and determined by real-time quantitative PCR.
Antibiotic susceptibility testing of T. whipplei strains
To be sure that the decrease in DNA copies when infected cells were treated by antibiotics was not due to a loss of viability, we have added in each PCR experiment controls, including the infected cells without antibiotics at day 0 and at day 12. Antibiotic susceptibilities against the three isolates by using real-time PCR were determined on day 12 because the three isolates reached their maximum growth after this time. This was confirmed by Gimenez staining. The three isolates of T. whipplei had a similar pattern of antibiotic susceptibility (Table 2). They were susceptible to doxycycline, macrolides, telithromycin, penicillin G, streptomycin, rifampin, teicoplanin, chloramphenicol, thiamphenicol, amoxicillin, and trimethoprim-sulfamethoxazole, with MICs ranging between 0.25 and 2 μg/ml. We found heterogeneity in the susceptibility to imipenem with the Twist strain being susceptible (MIC of 0.5 μg/ml), whereas Endo2 and Slow isolates were resistant (MIC of 10 μg/ml). To vancomycin the MICs were ≥10 μg/ml for the three strains. Antibiotics that were less active were cephalotin, colimycin, aztreonam, and the fluoroquinolones.
TABLE 2.
MICs of antibiotics active against T. whipplei as determined by Light Cycler assay
| Antibiotic | MICs (μg/ml)
|
||
|---|---|---|---|
| Twist | Endo-5 | Slow | |
| Penicillin G | 1 | 0.5 | 0.5 |
| Amoxicillin | 0.5 | 1 | 1 |
| Rifampin | 0.5 | 1 | 2 |
| Erythromycin | 1 | 1 | 2 |
| Clarithromycin | 2 | 2 | 2 |
| Telithromycin | 0.25 | 0.5 | 0.5 |
| Doxycycline | 1 | 2 | 2 |
| Sulfamethoxazole-trimethoprim | 2/0.5 | 4/1 | 4/1 |
| Teicoplanin | 0.25 | 0.5 | 0.5 |
| Vancomycin | 10 | 10 | 10 |
| Streptomycin | 0.5 | 1 | 1 |
| Gentamicin | 4 | 8 | 8 |
| Thiamphenicol | 0.25 | 0.25 | 1 |
| Chloramphenicol | 1 | 1 | 2 |
| Imipenem | 0.5 | 10 | 10 |
| Aztreonam | 10 | 10 | 20 |
| Cephalotin | 10 | 10 | 20 |
| Ceftriaxone | 10 | 10 | 10 |
| Colimycine | 20 | 20 | 40 |
| Levofloxacin | 0.25 | 0.5 | 0.5 |
| Ofloxacin | 4 | 4 | 4 |
| Ciprofloxacin | 4 | 4 | 4 |
| Hydroxychloroquine | 2 | 4 | 8 |
Antibiotic dilutions were found to be stable for 15 days and had the same activity against control strains over this time in the control experiments.
Bactericidal activity of combined therapy
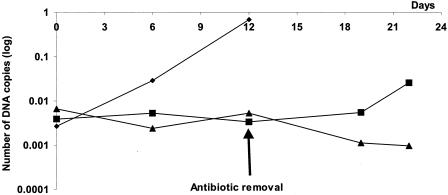
Cultures of Twist strain treated with doxycycline (2 μg/ml) alone or doxycycline (2 μg/ml) and hydroxychloroquine (1 μg/ml) for 12 days, were reinoculated onto fresh confluent cells without antibiotics to evaluate the regrowth of the bacteria. The results of this experiment are shown in Fig. 3. The combination of doxycycline and hydroxychloroquine appeared to be bactericidal since no bacterial regrowth was detected by Gimenez staining or PCR assay, even after 10 days of subculture. When used alone, doxycycline was not bactericidal since regrowth of organisms occurred after 10 days of subculture as detected by PCR assay (+1.65 log of DNA copies) and also by Gimenez staining.
FIG. 3.
Growth kinetics of T. whipplei Twist strain treated for 12 days with either doxycycline alone or doxycycline plus hydroxychloroquine. Antibiotics were removed after 12 days and samples were reinoculated in fresh confluent MRC5 cells. Regrowth of the bacteria was determined by quantitative PCR. Symbols: ♦, growth control; ▪, doxycycline (2 μg/ml); ▴, doxycycline (2 μg/ml) plus hydroxychloroquine (1 μg/ml).
DISCUSSION
Since T. whipplei are obligate intracellular gram-positive bacteria, cell culture systems are required for in vitro studies of antibiotic susceptibility. The phenotypic assays usually used to determine the antibiotic susceptibility of intracellular bacteria are not suitable for T. whipplei, and thus we recently developed a genotypic assay using the LightCycler instrument (18). This assay enabled us to demonstrate the natural resistance of T. whipplei to fluoroquinolones. The resistance is associated with mutations in the DNA gyrase gene that is the target for this class of antibiotic (18). In the present study, we used the same method to assess the activity of a wide range of antibiotics encompassing members of almost all of the available antibiotic classes. We confirmed the specificity of the primers we used for the ITS gene with our findings that the PCR products obtained were always 100% identical to those of T. whipplei. Growth kinetics of the three isolates were similar to those described in our previous report (18). Determination of MICs was carried out in duplicate in three different experiments and results were always consistent. Our assay was thus reproducible and enabled us to determine the MICs of antibiotics precisely. We report for the first time an extensive study on the susceptibility of T. whipplei in cell culture to antibiotics.
Although most antibiotics were active, doxycycline, cotrimoxazole, rifampin, penicillin, macrolides, and aminoglycosides were particularly active. These results, apart from those for the penicillins, are consistent with our current knowledge of antibiotic activity against intracellular bacteria (21). Indeed, doxycycline, rifampin, macrolides, and aminoglycosides have been shown to be highly active against strict intracellular bacteria such as Rickettsia spp., C. burnetii, and Ehrlichia spp. (21, 22, 32). Of note is our finding that trimethoprim-sulfamethoxazole was active. The target for trimethoprim is dihydrofolate reductase, the coding sequence of which has recently been shown to be absent from the genome of T. whipplei (3). The activity of trimethoprim-sulfamethoxazole in our study was therefore likely to be due to an effect of the sulfonamide compound alone. Dihydropteroate synthetase, the target gene for sulfamethoxazole, occurs in both the genome sequences available for T. whipplei. This activity is similar to that reported for bacteria of the genus Nocardia spp. (34). We found that penicillin compounds were active against T. whipplei, although this class of antibiotics usually have poor penetration into cells. Their activity could be due to the inhibition of growth of extracellular bacteria (15, 29, 30) since supernatants of cultures treated with penicillins were free of the extracellular form of the bacteria (data not shown).
The heterogeneity of susceptibility to imipenem that we found was very surprising because susceptibility to penicillin G and resistance to imipenem has not been described previously. Resistance to imipenem has been reported in other gram-positive bacteria, including Enterococcus, Nocardia, and Corynebacteria spp. (10). The mechanisms whereby T. whipplei are resistant to imipemen are unknown, but it could be due to alterations in penicillin-binding proteins, to the presence of zinc metalloprotease (beta-lactamase), to efflux mechanisms, or to intracellular inactivation of the drug (10).
Among glycopeptide compounds, although teicoplanin was active, we found a low level of resistance to vancomycin (MICs of 10 μg/ml). This discrepancy in activity has been reported previously for bacteria in the genus Enterococcus due to the acquisition of vanC or vanE operons, which code for ligases (2). The vanE operon, in particular, confers resistance to vancomycin, whereas teicoplanin and ampicillin remain active (2, 9). Of interest is our finding of a d-Ala-d-Ala ligase (GenBank accession number NP_789196) in the genome of T. whipplei, which has 32% homology with the vanE operon from E. faecalis (GenBank accession number AAL27442.1) and could be involved in the mechanism of resistance in T. whipplei.
Antibiotics found to be less active in our study were cephalotin, colimycin, aztreonam, and fluoroquinolones. This poor activity could be due either to a poor penetration in the cells, to an intracellular interaction, or to efflux mechanisms. The clear explanation of this poorer activity is not known except for the fluoroquinolones, for which there are specific mutations within the DNA gyrase gene involved in resistance (18). Our results are in accordance with our recent report showing that these antibiotics may be used to decontaminate samples from which T. whipplei is to be isolated in cell culture (6).
Based on our knowledge of the activity of antibiotics against C. burnetii, a bacterium that also exists in intracellular acidic vacuoles (12), we tested the in vitro bactericidal effect of a combination of doxycycline and hydroxychloroquine on the organism. Alkalinization of the vacuoles has been shown to result in a bactericidal activity of doxycycline against C. burnetii (19, 25) and S. aureus (20). We also found that this also occurred with T. whipplei and that the bacteria were killed when a combination of doxycycline and hydroxychloroquine were used. This finding could help to better define the optimum treatment of Whipple's disease. Most patients suffering from chronic Whipple's disease may have their disease controlled by various antibiotic regimens, but the disease often recurs after antibiotic withdrawal. We believed that these treatment failures are due to a lack of bactericidal activity of the antibiotics used in therapy. This has been also demonstrated in patients with chronic Q fever (26). Because C. burnetii and T. whipplei both occur in acidic vacuoles of the cell during chronic infections, we suggest that our knowledge of the treatment of Q fever should be applied to Whipple's disease. Clinical multicentric trials using combination therapy with doxycycline and hydroxychloroquine as the main treatment of Whipple's disease will be needed to prove our hypothesis.
Acknowledgments
This study was supported in part by a grant from Aventis, Paris, France.
We thank Pat Kelly for reviewing the manuscript and for English language corrections.
REFERENCES
- 1.Bentley, S. D., M. Maiwald, L. D. Murphy, M. J. Pallen, C. A. Yeats, L. G. Dover, H. T. Norbertczak, G. S. Besra, M. A. Quail, D. E. Harris, A. Von Herbay, A. Goble, S. Rutter, R. Squares, S. Squares, B. G. Barrell, J. Parkhill, and D. A. Relman. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637-644. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D. A., T. Cabral, P. Van Caeseele, J. Wylie, and M. R. Mulvey. 2002. Molecular characterization of the vanE gene cluster in vancomycin-resistant Enterococcus faecalis N00-410 isolated in Canada. Antimicrob. Agents Chemother. 46:1977-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon, R. 2003. Whipple's disease, genomics, and drug therapy. Lancet 361:1916. [DOI] [PubMed] [Google Scholar]
- 4.Dobbins, W. O. 1995. Whipple's disease, p. 1030-1032. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and pratice of infectious diseases. Churchill Livingstone, New York, N.Y.
- 5.Dykman, D. D., B. A. Cuccherini, I. J. Fuss, L. W. Lum, J. E. Woodward, and W. Strober. 1999. Whipple's disease in a father-daughter pair. Dig. Dis. Sci. 44:2542-2544. [DOI] [PubMed] [Google Scholar]
- 6.Fenollar, F., M. L. Birg, V. Gauduchon, and D. Raoult. 2003. Culture of Tropheryma whipplei from human samples: a 3-year experience (1999 to 2002). J. Clin. Microbiol. 41:3816-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenollar, F., P. E. Fournier, D. Raoult, R. Gerolami, H. Lepidi, and C. Poyart. 2002. Quantitative detection of Tropheryma whipplei DNA by real-time PCR. J. Clin. Microbiol. 40:1119-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenollar, F., and D. Raoult. 2001. Whipple's disease. Clin. Diagn. Lab. Immunol. 8:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fines, M., B. Perichon, P. Reynolds, D. F. Sahm, and P. Courvalin. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fish, D. N. 1998. Carbapenems, p. 690-703. In V. L. Yu, T. C. Merigan, Jr., and S. L. Barriere (ed.), Antimicrobial therapy and vaccines. The Williams & Wilkins Co., Baltimore, Md.
- 11.Fleming, J. L., R. H. Wiesner, and R. G. Shorter. 1988. Whipple's disease: clinical, biochemical, and histopathologic features and assessment of treatment in 29 patients. Mayo Clin. Proc. 63:539-551. [DOI] [PubMed] [Google Scholar]
- 12.Ghigo, E., C. Capo, M. Aurouze, C. H. Tung, J. P. Gorvel, D. Raoult, and J. L. Mege. 2002. Survival of Tropheryma whipplei, the agent of Whipple's disease, requires phagosome acidification. Infect. Immun. 70:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimenez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 14.Khan, M. A., C. Herzog, J. St Peter, G. Hartley, R. Madlon-Kay, C. Dick, R. Asinger, and J. Vessey. 1998. The prevalence of cardiac valvular insufficiency assessed by transthoracic echocardiography in obese patients treated with appetite-suppressent drugs. N. Engl. J. Med. 339:765-766. [DOI] [PubMed] [Google Scholar]
- 15.La Scola, B., F. Fenollar, P. E. Fournier, M. Altwegg, M. N. Mallet, and D. Raoult. 2001. Description of Tropheryma whipplei gen. nov., sp. nov., the Whipple's disease bacillus. Int. J. Syst. Evol. Microbiol. 51:1471-1479. [DOI] [PubMed] [Google Scholar]
- 16.Maiwald, M., H. J. Ditton, A. Von Herbay, F. Rainey, and E. Stackebrandt. 1996. Reassessment of the phylogenetic position of the bacterium associated with Whipple's disease and determination of the 16S-23S ribosomal intergenic spacer sequence. Int. J. Syst. Bacteriol. 46:1078-1082. [DOI] [PubMed] [Google Scholar]
- 17.Maizel, H., J. Ruffin, and W. Dobbins. 1970. Whipple's disease: a review of 19 patients from one hospital and a review of the literature since 1950. Medicine 49:175-205. [PubMed] [Google Scholar]
- 18.Masselot, F., A. Boulos, M. Maurin, J. M. Rolain, and D. Raoult. 2003. Molecular evaluation of antibiotic susceptibility, the Tropheryma whipplei paradigm. Antimicrob. Agents Chemother. 47:1658-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurin, M., A. M. Benoliel, P. Bongrand, and D. Raoult. 1992. Phagolysosomes of Coxiella burnetii-infected cell lines maintain an acidic pH during persistent infection. Infect. immun. 60:5013-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurin, M., and D. Raoult. 1994. Phagolysosomal alkalinization and intracellular killing of Staphylococcus aureus by amikacin. J. Infect. Dis. 169:330-336. [DOI] [PubMed] [Google Scholar]
- 21.Maurin, M., and D. Raoult. 1996. Optimum treatment of intracellular infection. Drugs 52:45-59. [DOI] [PubMed] [Google Scholar]
- 22.Maurin, M., and D. Raoult. 2001. Use of aminoglycosides in treatment of infections due to intracellular bacteria. Antimicrob. Agents Chemother. 45:2977-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naegeli, B., F. Bannwart, and O. Bertel. 2000. An uncommon cause of recurrent strokes Tropheryma whippelii endocarditis. Strokes 31:2002-2003. [DOI] [PubMed] [Google Scholar]
- 24.Raoult, D., M. L. Birg, B. La Scola, P. E. Fournier, M. Enea, H. Lepidi, V. Roux, J. C. Piette, F. Vandenesch, D. Vital Durand, and T. J. Marrie. 2000. Cultivation of the bacillus of Whipple's disease. N. Engl. J. Med. 342:620-625. [DOI] [PubMed] [Google Scholar]
- 25.Raoult, D., M. Drancourt, and G. Vestris. 1990. Bactericidal effect of doxycycline associated with lysosomotropic agents on Coxiella burnetii in P388D1 cells. Antimicrob. Agents Chemother. 34:1512-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raoult, D., P. Houpikian, H. Tissot Dupont, J. M. Riss, J. Arditi-Djiane, and P. Brouqui. 1999. Treatment of Q fever endocarditis: comparison of two regimens containing doxycycline and ofloxacin or hydroxychloroquine. Arch. Int. Med. 159:167-173. [DOI] [PubMed] [Google Scholar]
- 27.Raoult, D., H. Ogata, S. Audic, C. Robert, K. Suhre, M. Drancourt, and J. M. Claverie. 2003. Tropheryma whipplei Twist: a human pathogenic actinobacteria with a reduced genome. Genome Res. 13:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratliff, N., J. McMahon, T. Naab, and D. Cosgrove. 1984. Whipple's disease in the porcine leaflets of a carpentier-edwards prosthetic mitral valve. N. Engl. J. Med. 311:902-903. [DOI] [PubMed] [Google Scholar]
- 29.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 30.Silva, M. T., P. M. Macedo, and J. F. Moura Nunes. 1985. Ultrastructure of bacilli and the bacillary origin of the macrophagic inclusions in Whipple's disease. J. Gen. Microbiol. 131(Pt. 5):1001-1013. [DOI] [PubMed] [Google Scholar]
- 31.Singer, R. 1998. Diagnosis and treatment of Whipple's disease. Drugs 55:699-704. [DOI] [PubMed] [Google Scholar]
- 32.Spicer, A. J., M. G. Peacock, and J. C. Williams. 1981. Effectiveness of several antibiotics in suppressing chick embryo lethality during experimental infections by Coxiella burnetii, Rickettsia typhi, and R. rickettsii, p. 375-383. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, Inc., New York, N.Y.
- 33.Vital-Durand, D., C. Lecomte, P. Cathebras, H. Rousset, and P. Godeau. 1997. Whipple disease: a clinical review of 52 cases. Medicine 76:170-184. [DOI] [PubMed] [Google Scholar]
- 34.Wallace, R. J., Jr., L. C. Steele, G. Sumter, and J. M. Smith. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob. Agents Chemother. 32:1776-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whipple, G. H. 1907. A hitherto undescribed disease characterized anatom ically by deposits of fat and fatty acids in the intestinal and mesenteric lymphatic tissues. Bull. Johns Hopkins Hosp. 18:382-393. [Google Scholar]