Abstract
MetaboLights (http://www.ebi.ac.uk/metabolights) is the first general-purpose, open-access repository for metabolomics studies, their raw experimental data and associated metadata, maintained by one of the major open-access data providers in molecular biology. Metabolomic profiling is an important tool for research into biological functioning and into the systemic perturbations caused by diseases, diet and the environment. The effectiveness of such methods depends on the availability of public open data across a broad range of experimental methods and conditions. The MetaboLights repository, powered by the open source ISA framework, is cross-species and cross-technique. It will cover metabolite structures and their reference spectra as well as their biological roles, locations, concentrations and raw data from metabolic experiments. Studies automatically receive a stable unique accession number that can be used as a publication reference (e.g. MTBLS1). At present, the repository includes 15 submitted studies, encompassing 93 protocols for 714 assays, and span over 8 different species including human, Caenorhabditis elegans, Mus musculus and Arabidopsis thaliana. Eight hundred twenty-seven of the metabolites identified in these studies have been mapped to ChEBI. These studies cover a variety of techniques, including NMR spectroscopy and mass spectrometry.
INTRODUCTION
Metabolomics is the systematic study of the small molecular metabolites in a cell, tissue, biofluid or cell culture media that are the tangible result of cellular processes or responses to an environmental stress (1,2). The identification and quantification of such metabolites provide unique insights into the metabolic processes that are taking place in the cellular environment. Metabolic profiles taken from body fluids have the potential to act as biomarkers for many different diseases, an approach that has already shown value in, for example, heart disease and diabetes (3), the effects of diet (4) and interactions with the environment (5). Metabolomics technologies yield many insights into basic biological research in areas such as systems biology and metabolic modeling (6), pharmaceutical research (7), nutrition (8) and toxicology (9). However, to harness the full potential of metabolomics, researchers needs access to data and knowledge to compare, contrast and make inferences from the results they obtain in their experiments (10). The metabolome is the total complement of metabolites present in a biological sample under given genetic, nutritional or environmental conditions. Since such conditions can vary dramatically, it is clear that databases will need to collect numerous experiments together for a given species to accurately reflect the underlying diversity and complexity. In recent years, several instrument or species-specific dedicated metabolomics reference databases have been created. Examples include the Human Metabolome Database [HMDB, http://www.hmdb.ca, (11)], the Biological Magnetic Resonance Data Bank [BMRB, http://www.bmrb.wisc.edu, (12)], METLIN [http://metlin.scripps.edu, (13)], LIPIDMAPS [http://www.lipidmaps.org, (14)] and more general databases such as KNApSAck (http://kanaya.aist-nara.ac.jp/KNApSAcK/). However, the various metabolomics communities worldwide have not had a global open repository to share experimental data and associated metadata across species and platforms. MetaboLights will (i) provide a single point of access to worldwide data and knowledge in metabolomics, (ii) facilitate the development and adoption of a common data sharing format, (iii) ensure data traceability and reproducibility and (iv) progressively promote interoperability across existing resources.
MetaboLights consists of two distinct layers: a repository, enabling the metabolomics community to share findings, data and protocols for any form of metabolomics study, and a reference layer of curated knowledge about metabolites (forthcoming). MetaboLights is not intended to replace specialist resources but is specifically designed to build on prior art and extensively collaborate with the existing databases to ensure that data are exchanged and that assimilation efforts target gaps in worldwide available knowledge. We are dedicated to close collaboration with all major parties involved in the creation of this prior art, such as the Metabolomics Society, Metabomeeting and the Metabolomics Standards Initiative (MSI) (15). MetaboLights is working towards the setup of formal data sharing agreements with major resources such as the HMDB, the Golm Metabolome Database (16), MetabolomeExpress (17) and the Riken Metabolomics Platform (18). MetaboLights contains references to identified metabolites in existing databases, such as HMDB and ChEBI (19), and does not duplicate compound information residing in these external databases. Rather, it uses programmatic access to retrieve relevant data to display a unified metabolite-centric view to our users. In the future, such metabolite-centric views will be extended to show metabolites in the context of pathways, harnessing the Reactome database of biochemical pathways (20). In this article, we report on the structure and content of the MetaboLights repository and describe on-going work in the development of the reference layer.
DATABASE DESCRIPTION
The MetaboLights repository can be accessed at http://www.ebi.ac.uk/metabolights and http://metabolights.org.
Database content
We store and display an extensive set of associated information for studies in MetaboLights. This includes submitter and author information, publication references, the study design, protocols applied, names of data files included, platform information and metabolite information. The metabolite information includes a description, external database identifiers, formula and intensity or concentration, and where the metabolite was identified in the sample.
At present, the repository includes 15 submitted studies, of which 10 are publicly visible. These studies encompass 93 protocols for 714 assays, and span over 8 different types of organism including human, Caenorhabditis elegans, Mus musculus and Arabidopsis thaliana. Eight hundred twenty-seven of the metabolites identified in these studies have been mapped to ChEBI and 136 to HMDB. Thirty-eight users are currently registered.
Technical architecture
The MetaboLights repository is based on open source freely available software and tools. The web application runs on an Apache Tomcat server and the database backend is an Oracle database, but other standard SQL databases like MySQL and PostgreSQL can be used.
At the core of the database implementation is the ISA framework (21). The main database schema is powered by the ISA BioInvestigation Index (BII), which contains user information and all searchable metadata for the studies. Currently, there are 72 tables in this database schema. Any data-files that are associated with a study are stored on a traditional file system, and only their reference is stored in the database. Each study has a separate folder on the file systems containing the study metadata and associated files. This ensures a relative small database schema, but individual studies can be very large depending on the size of attached data files.
Searching for data
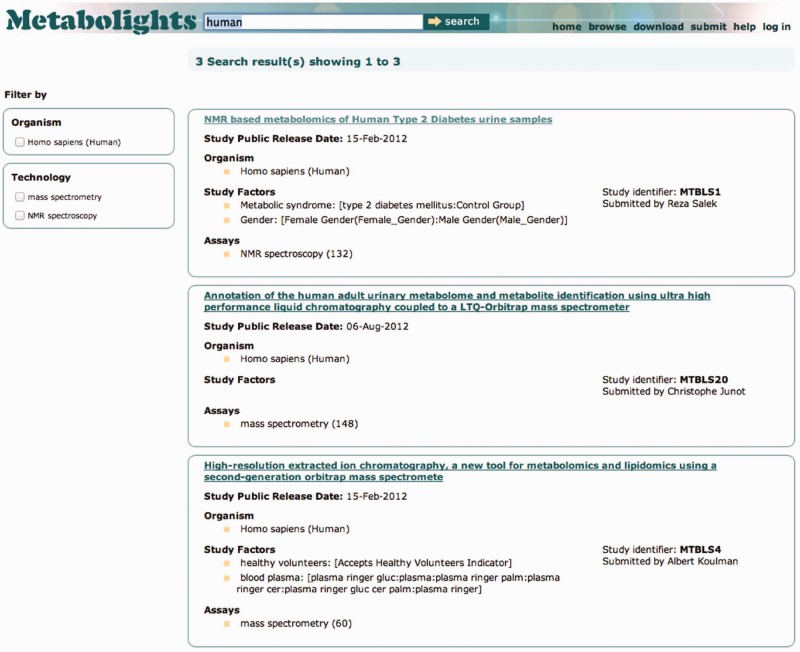
The online search facility provides the ability to search using free text through most of the underlying data fields, including the study description, study title, protocols, metabolites and authors. Currently, we support free-text searching and you can combine multiple search terms, for example ‘human urine’ will give you all studies where you find the terms ‘human’ and ‘urine’ are used. The search result page, as illustrated in Figure 1, shows general study information like the submitter of the study, the study title, public release date, organism(s), study design and platform.
Figure 1.
Searching for ‘human’ in the MetaboLights web application.
It is possible to further refine the search result using ‘search facets’. Search facets give the user the ability to limit the search results to a selection of species, platform and metabolite. For example, if you select a specific organism from the filter, the search results are limited to show only studies containing this organism. The search mechanism in MetaboLights is implemented using a text index (Lucene index) so no direct backend database queries are performed during a general search. This ensures a fast search facility.
Figure 1 shows the search results page when searching for ‘human’ across all of MetaboLights. To see the details of a study, the user can simply click on the study title. Example of what is displayed in the study details are in Supplementary Figures S1–S4. These images show screenshots of the web interface of MetaboLights with study data loaded for an NMR-based metabolomics study, MTBLS1. The Study details page consists of four tabs. The first tab (Supplementary Figure S1) shows information about the submitters, the relevant dates, study title and description, organisms, study design, publications and the experimental factors. The next tab (Supplementary Figure S2) details the protocols used during this study, from how the sample was collected through to the metabolite identification. Next, we have the data tab (Supplementary Figure S3). Here, we show data files for this study, detailed for technology platform used and experimental factors. Finally, we have metabolite identification (Supplementary Figure S4). Each identified metabolite has an external database reference, for example a ChEBI or HMDB identifier. Metabolites identified with a ChEBI accession show additional molecule description. The identified metabolite tab details which sample the compound was identified in. Unknown compounds are listed without a database reference.
Browsing data
Users can browse studies in MetaboLights using the ‘browse’ link. This will give a complete list of all the public studies currently available. If the user is registered and currently logged in to MetaboLights, additional private studies may be displayed. These private studies are either under the users control or have been directly shared from other users. To limit the number of studies in the browsing list, the user can activate the same facets available for a general search.
Downloads and programmatic access
MetaboLights software components are open source and all data are free to download and use for any purpose. All public studies are downloadable as ISA-Tab (22) metadata files with associated data files directly from the online study details page, and from the MetaboLights download page http://www.ebi.ac.uk/metabolights/download. A direct bulk download using ftp is available from ftp://ftp.ebi.ac.uk/pub/databases/metabolights/, organized into sub-folders for public studies. There are no web services for programmatic access available at present. However, this functionality is scheduled for a future release of the repository.
Submitting data
MetaboLights accepts experimental descriptions in ISA-Tab format, which can be created by the ISAcreator editor tool. MetaboLights also offers different templates for the ISAcreator tool to accommodate the description of different types of metabolomics experiments. ISAcreator is a standalone Java desktop application that enables researchers to report experimental information, associate raw and processed data files, and submit the collated information to the MetaboLights database. Building on the OSGI plugin architecture, the ISAcreator has been extended to create a ‘Metabolite Identification’ add-on to capture relevant information for all small molecules identified in a study, with a link to a relevant chemical database (Figure 2). MetaboLights also accepts studies that have unknown or incomplete metabolite identification. This information has the potential to facilitate the identification of unknown metabolites in the future.
Figure 2.
Part of Study MTBLS1 in ISAcreator with the Metabolite Identification Plugin active.
Currently, we accept all data formats for ‘raw’ instrumental data, converted open source file formats and any processed data, but we strongly recommend that processed data should be made available in open formats, such as mzML (23) for MS data.
MetaboLights implements metadata guidelines according to the recommendations of the Metabolomics Standards Initiative (MSI). The MSI defined a set of metabolomics reporting standards by harnessing and coordinating the efforts of several pre-existing international initiatives. MSI developed checklists and standards that have subsequently been adopted by the community, including minimum metadata reporting recommendations (24).
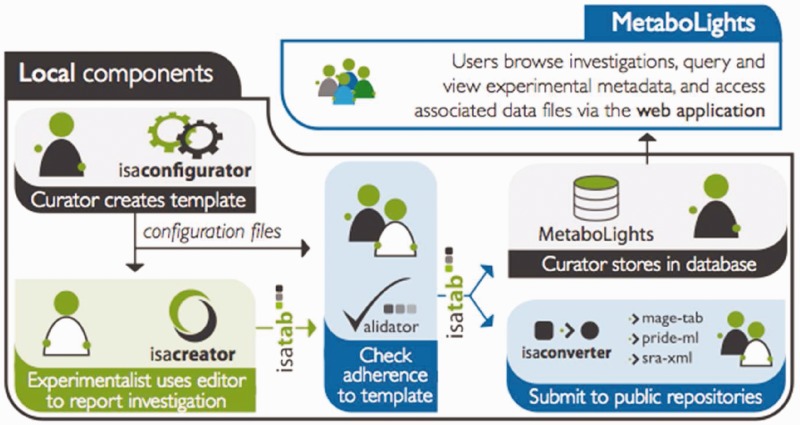
To facilitate high quality data submissions for NMR or MS experiments, there is a guided submission process to help meet MSI recommendations and extensively use community-developed controlled vocabularies and ontologies. ISAcreator also provides advanced mechanisms for mapping to and uploading information from existing spreadsheets. Figure 3 illustrates the ISA components in a typical data creation scenario.
Figure 3.
A typical workflow, using the ISA framework, for reporting information and submitting it to the MetaboLights database.
An R package has been developed to facilitate data analysis (Supplementary Method). The Risa module, available in the next BioConductor release, includes functionality to process mass spectrometry data relying on the xcms package (25), and to save analysis results back to ISA archives.
Installing a local copy of the MetaboLights repository
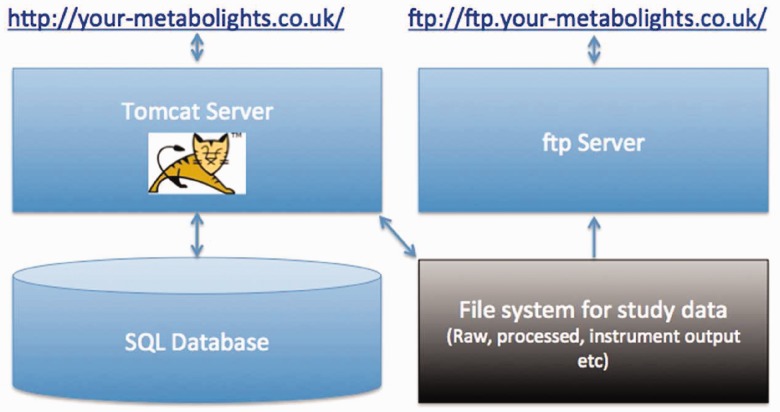
To install MetaboLights locally, you require a SQL database (MySQL, PostgreSQL or Oracle), a subversion client (svn) and an Apache Tomcat server. The MetaboLights Repository source code can be found at http://sourceforge.net/projects/metabolomes, here you will also find more details regarding how to install MetaboLights locally. The ISAcreator Metabolite Identification plugin can be found at: https://github.com/EBI-Metabolights/ISAcreatorPlugins. The ISA framework is also open source and is available at: https://github.com/ISA-tools. Figure 4 shows the principal components of a local MetaboLights repository installation.
Figure 4.
Simple technical architecture of a local MetaboLights repository, including web (http) and ftp access.
Access and privacy policy
MetaboLights grants free access and reuse of the public data it stores to everyone. Only registered users can upload and share study data. To facilitate deposition of research data not yet publicly visible, the submitter can set a data embargo for a period of up to 60 months, which can be lifted on results publication or extended upon request. Submitters can also request for access to their private data to be granted to specific other registered users. This feature may be particularly useful in facilitating collaborations and the peer review process.
Feedback
To facilitate user feedback, we have created a SourceForge tracker for logging issues, available at http://sourceforge.net/projects/metabolomes. There is also an online contact form, http://www.ebi.ac.uk/metabolights/contact, and contact email address, metabolights-help@ebi.ac.uk.
DISCUSSION
The MetaboLights repository was launched on the 28 June 2012 at the 8th International Conference of the Metabolomics Society in Washington, DC, USA. The repository is now accepting study submissions from a growing number of active users worldwide with submission privileges. For the latest statistics on current studies and submitters, please see http://www.ebi.ac.uk/metabolights/stats.
The requirement by a growing number of publishers and funding agencies to deposit data associated with journal publications to public repositories is expected to motivate a substantial number of future submissions. As more datasets become available, Metabolights will become an invaluable resource for those wishing to develop new algorithms for the processing of metabolomic data. The creation of a long-term institution-backed, as it will be maintained by EBI after the grant ends, public repository such as MetaboLights at EMBL-EBI allows laboratories across the globe to collaborate on projects through data sharing, and thereby to begin to collaboratively generate the large datasets needed to address how the environment, genome and diet influence the metabolome of a species.
Future work
The MetaboLights team is now actively specifying the MetaboLights Reference Layer, which will be launched in Summer 2013. The Reference Layer will be a comprehensive knowledge base organized around a metabolite-centric view, and will include elements such as reference spectra of various types, biological reference data, protocols, cross-references to other resources and advanced search and download functionality. There will be comprehensive manually curated data, including chemical structures and characteristics from ChEBI, metabolic pathways, reference spectroscopy and chromatography. Furthermore, there will be information about the reference biology, metabolites and their occurrence and concentration in species, organs, tissues and cellular compartments in various conditions, both healthy and diseased. Publication references and protocols will also be available. This will enable experimentalists to get a comprehensive Metabolomic view on known metabolites.
We are also substantially enhancing our online help capabilities with online video instructions as well as detailed scenarios for completing new studies for submission. A new section with ‘Gold Standard Studies’ will be included for easy reference. These studies can be used as templates for similar experiments.
In October 2012, the European COordination of Standards in MetabOlomicS (COSMOS) consortium, comprising 14 European partners, will start its work on Metabolomics data standardization, publication and dissemination workflows. The MetaboLights database is a key component in this effort. It is the aim of the COSMOS project to develop efficient policies to ensure that Metabolomics data are encoded in open standards, tagged with a community-agreed and complete set of metadata, supported by a communally developed set of open source data management and capturing tools, disseminated in open-access databases adhering to these standards, supported by vendors and publishers, who require deposition upon publication, and properly interfaced with data in other biomedical and life science e-infrastructures [such as ELIXIR (26), BioMedBridges (http://www.biomedbridges.eu), EU-OPENSCREEN (http://www.eu-openscreen.de) and BBMRI (http://bbmri.eu)].
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4 and Supplementary Methods.
FUNDING
The development of MetaboLights is funded by the BBSRC [BB/I000933/1]; ISA framework by the BBSRC [BB/I025840/1, BB/I000771/1, BB/J020265/1 to S.A.S.]; NERC, EU [EC 312941 to S.A.S.]; University of Oxford e-Research Centre [to S.A.S.]. COSMOS is funded by European Commission Grant [EC312941]. Funding for open access charge: BBSRC [BB/1000933/1].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The MetaboLights project team thanks the following persons for their invaluable contributions: Rafael Alcántara, Masanori Arita, Mike Beale, Nick Bond, Kees van Bochove, Jildau Bouwman, Scarlet Brockmoeller, Steve Bryant, Hong Cao, Juan Castrillo, Jenny Cham, Cecilia Castro, Yajing Chu, Tim Ebbels, Michael Eiden, Oliver Fiehn, Andrew Gibbs, Roy Goodacre, Martin Hornshaw, Jan Hummel, Albert Koulman, Peter Meadows, Pablo Moreno, Theo Reijmers, Francis Rowland, Linda Scoriels, Mark Seymour, Tim Smith, Anthony Taylor, Chris Taylor, Michael Wakelam, Jane Ward and David Wishart.
REFERENCES
- 1.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 2.German JB, Hammock BD, Watkins SM. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics. 2005;1:3–9. doi: 10.1007/s11306-005-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson H. Meet the human metabolome. Nature. 2007;446:8. doi: 10.1038/446008a. [DOI] [PubMed] [Google Scholar]
- 4.Cheng KK, Benson GM, Grimsditch DC, Reid DG, Connor SC, Griffin JL. Metabolomic study of the LDL receptor null mouse fed a high-fat diet reveals profound perturbations in choline metabolism that are shared with ApoE null mice. Physiol. Genomics. 2010;41:224–231. doi: 10.1152/physiolgenomics.00188.2009. [DOI] [PubMed] [Google Scholar]
- 5.Veldhoen N, Ikonomou MG, Helbing CC. Molecular profiling of marine fauna: integration of omics with environmental assessment of the world’s oceans. Ecotoxicol. Environ. Saf. 2012;76:23–38. doi: 10.1016/j.ecoenv.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Kell DB. Metabolomics and systems biology: making sense of the soup. Curr. Opin. Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Xu EY, Schaefer WH, Xu QW. Metabolomics in pharmaceutical research and development: metabolites, mechanisms and pathways. Curr. Opin. Drug Discovery Dev. 2009;12:40–52. [PubMed] [Google Scholar]
- 8.Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am. J. Clin. Nutrition. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- 9.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 10.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 14.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Murphy RC, Raetz CRH, Russell DW, et al. Lmsd: lipid maps structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sansone SA, Fan T, Goodacre R, Griffin JL, Hardy NW, Kaddurah-Daouk R, Kristal BS, Lindon J, Mendes P, Morrison N, et al. The metabolomics standards initiative. Nat. Biotechnol. 2007;25:846–848. doi: 10.1038/nbt0807-846b. [DOI] [PubMed] [Google Scholar]
- 16.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al. Gmd@Csb.Db: the Golm metabolome database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 17.Carroll AJ, Badger MR, Harvey Millar A. The MetabolomeExpress Project: enabling web-based processing, analysis and transparent dissemination of GC/MS metabolomics datasets. BMC Bioinformatics. 2010;11:376. doi: 10.1186/1471-2105-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, Hirai MY, Sakurai T, Kikuchi J, Saito K. PRIMe: a web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008;8:339–345. [PubMed] [Google Scholar]
- 19.de Matos P, Alcantara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sansone SA, Rocca-Serra P, Field D, Maguire E, Taylor P, Hofmann O, Fang H, Neumann S, Tong W, Amaral-Zettler L, et al. Toward interoperable bioscience data. Nat. Genet. 2012;44:121–126. doi: 10.1038/ng.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocca-Serra P, Brandizi M, Maguire E, Sklyar N, Taylor C, Begley K, Field D, Harris S, Hide W, Hofmann O, et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26:2354–2356. doi: 10.1093/bioinformatics/btq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Rompp A, Neumann S, Pizarro AD, et al. mzML—a community standard for mass spectrometry data. Mol. Cell. Proteomics. 2011;10:R110.000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodacre R, Broadhurst D, Smilde AK, Kristal BS, Baker JD, Beger R, Bessant C, Connor S, Calmani G, Craig A, et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics. 2007;3:231–241. [Google Scholar]
- 25.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 26.Crosswell LC, Thornton JM. ELIXIR: a distributed infrastructure for European biological data. Trends Biotechnol. 2012;30:241–242. doi: 10.1016/j.tibtech.2012.02.002. [DOI] [PubMed] [Google Scholar]