Abstract
The Plant Resistance Genes database (PRGdb; http://prgdb.org) is a comprehensive resource on resistance genes (R-genes), a major class of genes in plant genomes that convey disease resistance against pathogens. Initiated in 2009, the database has grown more than 6-fold to recently include annotation derived from recent plant genome sequencing projects. Release 2.0 currently hosts useful biological information on a set of 112 known and 104 310 putative R-genes present in 233 plant species and conferring resistance to 122 different pathogens. Moreover, the website has been completely redesigned with the implementation of Semantic MediaWiki technologies, which makes our repository freely accessed and easily edited by any scientists. To this purpose, we encourage plant biologist experts to join our annotation effort and share their knowledge on resistance-gene biology with the rest of the scientific community.
INTRODUCTION
Plants protect themselves from disease by activating a broad array of defense responses that ultimately inhibit growth and spread of invading pathogens. The principal immune mechanism against pathogens in plants is mediated by resistance (R) gene (1). However, the essential components of this defense system remain still unknown even if it has been deeply investigated. High-throughput genomic experiments and plant genome sequences available in public databases are offering unprecedented opportunities to identify novel R-genes, to explore their function and their diversification process, to discover new resistance capacity, and ultimately to elucidate their mechanisms of interaction between pathogens and their plant hosts.
With the intention to facilitate research on this agriculturally important gene family, we launched in 2009 the Plant Disease Resistance Gene database, a comprehensive repository of R-genes across hundreds of plant species (2). PRGdb version 1.0 comprised a total of 16 844 gene entries, of which 73 were known and manual-curated R-genes (e.g. the ‘reference’ data set), 6308 were putative R-genes retrieved from NCBI Genbank, and 10 463 were putative R-genes computationally predicted from NCBI UniGene transcriptomic data using the in-house developed bioinformatics pipeline DRAGO (Disease Resistance Analysis and Gene Orthology) (2). In the last few years, several plant genome-sequencing projects have progressed rapidly. For instance, tomato (3), potato (4) and melon (5) genomes were recently completed, and thus providing the opportunity to discover additional R-genes. Therefore, the huge amount of sequence data not yet analysed for this gene family required a substantial update of our database for addressing the community requests.
In this manuscript, we present an update of the plant resistance gene database, PRGdb. In the release 2.0, our database has been expanded more than 6-fold to include useful biological information about a total of 104 459 R-genes (of which ∼90 000 newly annotated) from 233 plant species. Of these entries, 112 are manual-curated R-genes described in the literature to confer resistance to 122 different pathogens. All remaining genes were predicted by our DRAGO bioinformatics pipeline from sequences available in public transcriptomic and genomic databases (NCBI Genbank, NCBI UniGene, Phytozome) (6), as well as from gene models annotated in recent plant genome sequence projects, such as potato (4), tomato (3) and melon (5). Last but not least, we completely revised our web interface that is now based on Semantic MediaWiki technologies, which makes our repository freely accessible and easily editable by the scientific community. With this new wiki design, PRGdb aims to provide a forum for scientists working in this research field. Therefore, we do invite plant biology experts to join our efforts and help us on making PRGdb a more collaborative and comprehensive resource through their valuable contributions.
DATA UPDATE
To support the scientific community working on plant disease resistance area, we updated the PRGdb knowledge-base. In this release 2.0, the number of genes of previous version has grown more than 6-fold, new sections have been created and the domain analysis and the sequence classification of the candidate R-genes have been improved. To date, PRGdb 2.0 stores 112 ‘reference’ R-genes (40 more than the previous version), and 122 pathogen and 233 plant species (45 more than the previous version). The analysis with the DRAGO bioinformatics pipeline on 2012 NCBI ‘nr’ and UniGene databases yielded a total of 34 558 putative R-genes (20 000 more than the previous version). In addition to these traditional annotated categories, we included a new dataset predicted by DRAGO from genomic sequences available at Phytozome v8 (6), thus incrementing our repository of 68 509 putative R-genes from 31 plant sequenced genomes. Each putative R-gene sequence has been annotated with DNA and amino acid sequences, information about species and predicted protein domains. Noteworthy large-scale data released from PRGdb are listed in Table 1. Finally, another unique and important set of R-genes has been annotated by our team during the analysis of plant genome species recently sequenced and not yet present in the Phytozome database, such as tomato, potato and melon. For these species, a complete analysis of their R-genes panorama has been accomplished and a dedicated section of the database has been built to store, visualize, and consult these unique results (see wiki ‘metaspecies’ section). Table 2 shows a comparison of the R-gene classes identified in these agriculturally important species. Moreover, in this update, we include new identified R-protein domains, thus reaching a total of nine well-known domain types: the MLO (PFAM PF03094) (7), the RPW8 (8), and the GNK2 domain extracted from the antifungal protein Ginkobilobin2-1 (Interpro IPR002902) (9), and the already present CC (Coiled Coil), TIR (Toll-Interleukin like Region), NBS (Nucleotide Binding site), LRR (Leucine-Rich Region), KIN (Kinase) and Other (all other domains which have been described as conferring resistance through different molecular mechanisms) (2). The aforementioned new domain types helped to better classify the new candidate R-genes (Table 2). All data mentioned have been organized in new database sections as well-established repository for plant scientists. In particular, single gene pages are now available to directly visualize gene features allowing comparison of R-gene complements across entire plant lineages.
Table 1.
List of main entries present in PRG database
| Entries | No. |
|---|---|
| Plants | 233 |
| Pathogens | 122 |
| Domain types | 9 |
| Gene classes | 16 |
| Manually curated R-Genes | 112 |
| Putative R-Genes, collected from NCBI Protein | 9639 |
| Putative R-Genes, predicted from NCBI UniGene | 24 918 |
| Putative R-Genes, predicted from Pythozome | 68 498 |
| R-genes from plant genome sequencing projects | 1143 |
| Total number of plant genes | 104 310 |
| Avirulence genes | 23 |
| Diseases | 120 |
Table 2.
Putative R-genes retrieved in each sequenced species divided for class
| Species | CN | CNL | Mlo-like | N | NL | Other | RLK | RLK-GNK2 | RLP | RPW8-NL | T | TN | TNL | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aquilegia coerulea | 19 | 145 | 23 | 89 | 69 | 0 | 0 | 52 | 282 | 1 | 1 | 0 | 0 | 1010 | 1691 |
| Arabidopsis lyrata | 1 | 41 | 15 | 101 | 150 | 7 | 0 | 38 | 198 | 3 | 37 | 1 | 205 | 771 | 1568 |
| Arabidopsis lyrata subsp. lyrata | 3 | 36 | 23 | 41 | 40 | 0 | 0 | 15 | 142 | 1 | 7 | 0 | 25 | 629 | 962 |
| Arabidopsis thaliana | 65 | 748 | 40 | 125 | 230 | 46 | 53 | 90 | 580 | 19 | 180 | 12 | 683 | 3195 | 6066 |
| Areca catechu | 33 | 295 | 31 | 126 | 481 | 2 | 7 | 183 | 751 | 1 | 228 | 0 | 471 | 3217 | 5826 |
| Brachypodium distachyon | 4 | 89 | 6 | 15 | 57 | 1 | 0 | 22 | 70 | 0 | 1 | 0 | 0 | 240 | 505 |
| Brassica rapa | 31 | 71 | 21 | 65 | 40 | 10 | 2 | 72 | 306 | 3 | 26 | 0 | 119 | 1121 | 1887 |
| Capsella rubella | 18 | 66 | 16 | 47 | 25 | 12 | 1 | 63 | 242 | 5 | 43 | 0 | 56 | 785 | 1379 |
| Chlamydomonas reinhardtii | 3 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 165 | 193 |
| Citrus clementina | 18 | 340 | 24 | 101 | 176 | 1 | 15 | 28 | 449 | 1 | 15 | 0 | 84 | 1396 | 2648 |
| Citrus sinensis | 31 | 203 | 42 | 131 | 224 | 0 | 35 | 59 | 477 | 3 | 33 | 0 | 76 | 1916 | 3230 |
| Cucumis melo | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 5 |
| Cucumis sativus | 12 | 159 | 31 | 60 | 152 | 0 | 12 | 19 | 247 | 0 | 1 | 0 | 0 | 1047 | 1740 |
| Ectocarpus siliculosus | 0 | 0 | 2 | 15 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 118 | 136 |
| Eucalyptus grandis | 24 | 239 | 29 | 104 | 314 | 2 | 7 | 170 | 693 | 1 | 111 | 0 | 300 | 2332 | 4326 |
| Eutrema halophilum | 15 | 50 | 15 | 51 | 24 | 7 | 0 | 40 | 246 | 2 | 27 | 0 | 72 | 739 | 1288 |
| Glycine max | 37 | 132 | 60 | 190 | 281 | 0 | 8 | 141 | 803 | 8 | 78 | 0 | 229 | 3068 | 5035 |
| Gossypium arboreum | 7 | 170 | 20 | 56 | 75 | 0 | 0 | 36 | 232 | 1 | 1 | 0 | 0 | 827 | 1425 |
| Linum usitatissimum | 14 | 24 | 25 | 82 | 44 | 0 | 3 | 29 | 346 | 1 | 36 | 0 | 114 | 1227 | 1945 |
| Malus x domestica | 32 | 317 | 42 | 99 | 663 | 11 | 20 | 71 | 518 | 4 | 115 | 0 | 299 | 2811 | 5002 |
| Medicago truncatula | 15 | 98 | 29 | 105 | 357 | 0 | 3 | 58 | 297 | 4 | 95 | 0 | 291 | 1664 | 3016 |
| Oryza barthii | 0 | 12 | 15 | 71 | 100 | 0 | 0 | 19 | 195 | 1 | 9 | 0 | 33 | 721 | 1176 |
| Oryza sativa | 14 | 91 | 17 | 91 | 533 | 1 | 1 | 62 | 439 | 1 | 2 | 0 | 0 | 1548 | 2800 |
| Panicum virgatum | 37 | 332 | 24 | 115 | 532 | 0 | 2 | 64 | 478 | 0 | 3 | 0 | 0 | 2293 | 3880 |
| Pennisetum glaucum | 17 | 21 | 27 | 42 | 28 | 1 | 2 | 24 | 239 | 1 | 4 | 0 | 19 | 844 | 1269 |
| Phaseolus vulgaris | 18 | 114 | 29 | 69 | 227 | 0 | 3 | 67 | 306 | 2 | 18 | 0 | 114 | 1255 | 2222 |
| Physcomitrella patens | 7 | 6 | 21 | 89 | 38 | 1 | 0 | 0 | 225 | 1 | 2 | 0 | 6 | 770 | 1166 |
| Populus trichocarpa | 14 | 188 | 35 | 166 | 809 | 4 | 6 | 56 | 540 | 7 | 63 | 0 | 322 | 1946 | 4156 |
| Prunus persica | 8 | 132 | 22 | 69 | 138 | 0 | 0 | 30 | 273 | 1 | 19 | 0 | 131 | 981 | 1804 |
| Ricinus communis | 15 | 103 | 16 | 66 | 161 | 2 | 1 | 36 | 240 | 0 | 1 | 0 | 2 | 1224 | 1867 |
| Selaginella moellendorffii | 0 | 1 | 20 | 79 | 4 | 0 | 6 | 7 | 127 | 0 | 0 | 0 | 0 | 769 | 1013 |
| Setaria italica | 22 | 194 | 16 | 73 | 204 | 3 | 0 | 45 | 314 | 0 | 2 | 0 | 0 | 1143 | 2016 |
| Solanum lycopersicum | 2 | 26 | 8 | 9 | 41 | 1 | 0 | 4 | 45 | 0 | 1 | 0 | 3 | 407 | 547 |
| Solanum tuberosum | 0 | 7 | 5 | 4 | 71 | 0 | 2 | 3 | 5 | 0 | 8 | 0 | 4 | 288 | 397 |
| Sorghum bicolor | 5 | 13 | 16 | 94 | 320 | 3 | 0 | 45 | 298 | 1 | 5 | 0 | 0 | 1218 | 2018 |
| Vitis aestivalis | 14 | 102 | 23 | 61 | 98 | 0 | 8 | 25 | 261 | 1 | 13 | 0 | 29 | 1299 | 1934 |
| Vitis vinifera | 17 | 164 | 33 | 96 | 300 | 0 | 3 | 32 | 355 | 1 | 10 | 0 | 42 | 1268 | 2321 |
| Volvox carteri | 1 | 0 | 4 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 87 | 107 |
| Zea mays | 24 | 106 | 59 | 125 | 104 | 6 | 44 | 63 | 515 | 4 | 2 | 0 | 0 | 2330 | 3382 |
NEW FEATURES
Several scientific databases allow a direct interaction of users for sharing experimental and theoretical information. Most of them have a wiki structure and can contain specific or more generic data. Examples are Gene Wiki (10,11), WikiPathways (12), SubtiWiki (13), EcoliWiki (14), Tetrahymena genome database Wiki (15), Reactome (16) or SNPedia (17). Like these, PRGdb interface has been completely rewritten in a wiki system because in this way, it can be easier to get a larger scientific community be involved in the accumulation of data. We have used MediaWiki, the open-source software behind Wikipedia, to build this second version of the PRGdb with new features, a friendly interface, and new tools. Mapping our existing database structure into the wiki has been possible thanks to a set of Semantic MediaWiki extensions. This allows users to access and list different wiki pages according to multiple and flexible criteria and, at the same time, it offers a linking point for third parties to existing data via RDF triples attached to every page (18). By using Semantic technologies, PRGdb becomes another node of the linked data web (http://linkeddata.org/) On the other hand, it retrieves and links to resources such as Bio2RDF (19) (e.g. for retrieving additional species images) and potentially to other semantic empowered sites.
Other international plant databases use different models to interact with users. Among them, TAIR (20), SOLGenomics Network (21), Cucurbit Genomics Database (http://www.icugi.org) and Plant Organelles Database (22) are based on specific submission forms to contact database developers or systems based on information provided by emails; SOLGenomics Network has a forum section allowing users to interact as well. In our case, we take advantage of default wiki features such as change tracking and other social-like components, such as user pages, which show users activity and can be filled by plant disease resistance researchers. Moreover, apart from plain wiki editing for documentation purposes, several specific contribution entry points are also provided, so that registered users can suggest corrections or additions, such as related bibliography or even new reference genes. A group of reviewers, experts in the plant disease resistance field, will have the necessary permission to approve these contributions and, from that moment, these will be easily available and also linkable for third parties. In this way, PRGdb becomes a community database for plant scientists who could in turn contribute to this public resource collaboratively.
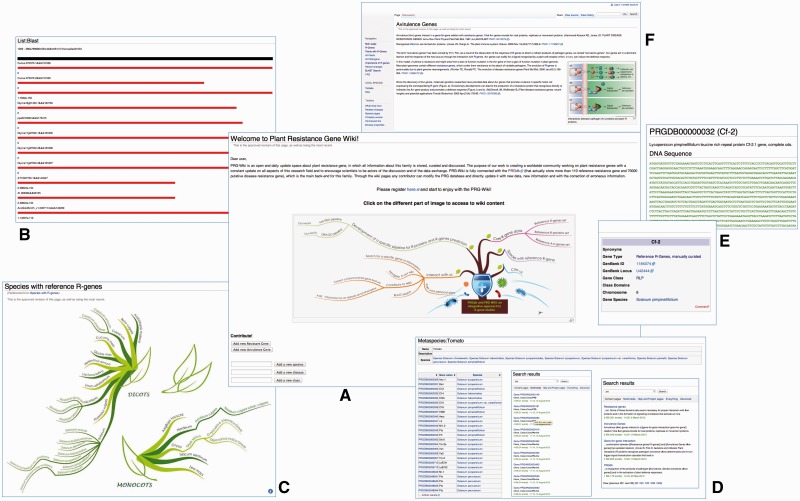
According with our final intent, wiki pages have been designed in a user-friendly version, enriched with interactive images and pages that summarize species with known R-genes (Figure 1). In the wiki pages, the user can find internal links connecting different sections of the website and improving accessibility to sequence, resistance gene data and related published information. New sections about plant and pathogens have been added, allowing users to quickly retrieve information from their species of interest. Some interactive images have also been included to facilitate navigation in the web site and to summarize results coming from data collection. Moreover, wiki pages are also intended to provide technical details about the pipelines adopted to predict R-genes and information coming from international scientific literature built collaboratively. In particular, notable scientist from all around the world that showed interest in our initiative will be asked to be in PRGdb review board. Reviewers will responsible for reviewing scientific accuracy of information. Members of primary specialty will be asked to review content areas that coincide directly with their expertise. Information in this section not only can be directly added by users, but can also be directly linked to other scientific databases. A variety of links to other database relevant to the topic are included in the initial menu.
Figure 1.
An overview of the wiki interface of the PRGdb version 2.0. (A) Home page; (B) BLAST results; (C) Clickable image of species annotated with R-genes; (D) Examples of metaspecies page, and two examples of wiki search results; (E) Reference R-gene wiki page; (F) Contributed wiki page.
Moreover, we have developed a new interface for BLAST search (23,24), implemented directly in the wiki system, so that results links are directly connected to gene wiki pages. All the sequences stored in the wiki are also provided as downloadable files, which are updated periodically from approved user wiki contributors.
FUTURE PERSPECTIVES
Newly sequenced plant genomes will be monitored from public resources, such as NCBI, Phytozome or any other specialized resource. Releases are expected to take place twice per year by integrating new data from our reference database plus the accumulated user contributions up to that moment. At the technical level, we will continue the employment of pipelines and algorithms to automate the management and classification of the sequence data and to better organize the content of the database. Connecting the various resources with phenotypic, genomic and molecular information will become a crucial task for mining results.
FUNDING
Italian Ministry of Education, University and Research (GenHort Project). Funding for open access charge: Department of Soil, Plant, Environment and Animal Production Sciences, University of Naples ‘Federico II’, via Università 100, 80055, Portici, Italy. Spanish Ministry of Economy and Competitiveness funded infrastructure technician support. Ref.: PTA2010-4446-I.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Valeria Morelli for the graphical support and the CRG staff for all the assistance provided to set up the web servers. Contribution no. 178 from the DISSPAPA.
REFERENCES
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Sanseverino W, Roma G, De Simone M, Faino L, Melito S, Stupka E, Frusciante L, Ercolano MR. PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res. 2010;38:D814–D821. doi: 10.1093/nar/gkp978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomato Genome C. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Potato Genome Sequencing Consortium, Xu,X., Pan,S., Cheng,S., Zhang,B., Mu,D., Ni,P., Zhang,G., Yang,S., Li,R. et al. (2011) Genome sequence and analysis of the tuber crop potato. Nature, 475, 189–195. [DOI] [PubMed]
- 5.Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, Gonzalez VM, Henaff E, Camara F, Cozzuto L, Lowy E, et al. The genome of melon (Cucumis melo L.) Proc. Natl. Acad. Sci. USA. 2012;109:11872–11877. doi: 10.1073/pnas.1205415109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 8.Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- 9.Miyakawa T, Miyazono K, Sawano Y, Hatano K, Tanokura M. Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins. 2009;77:247–251. doi: 10.1002/prot.22494. [DOI] [PubMed] [Google Scholar]
- 10.Good BM, Clarke EL, de Alfaro L, Su AI. The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res. 2012;40:D1255–D1261. doi: 10.1093/nar/gkr925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huss JW, 3rd, Lindenbaum P, Martone M, Roberts D, Pizarro A, Valafar F, Hogenesch JB, Su AI. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2010;38:D633–D639. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mader U, Schmeisky AG, Florez LA, Stulke J. SubtiWiki—a comprehensive community resource for the model organism Bacillus subtilis . Nucleic Acids Res. 2012;40:D1278–D1287. doi: 10.1093/nar/gkr923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntosh BK, Renfro DP, Knapp GS, Lairikyengbam CR, Liles NM, Niu L, Supak AM, Venkatraman A, Zweifel AE, Siegele DA, et al. EcoliWiki: a wiki-based community resource for Escherichia coli . Nucleic Acids Res. 2012;40:D1270–D1277. doi: 10.1093/nar/gkr880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stover NA, Punia RS, Bowen MS, Dolins SB, Clark TG. Tetrahymena genome database wiki: a community-maintained model organism database. Database. 2012;2012:bas007. doi: 10.1093/database/bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012;40:D1308–D1312. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Page RD. Linking NCBI to Wikipedia: a wiki-based approach. PLoS Curr. 2011;3:RRN1228. doi: 10.1371/currents.RRN1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belleau F, Nolin MA, Tourigny N, Rigault P, Morissette J. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J. Biomed. Inform. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mano S, Miwa T, Nishikawa S, Mimura T, Nishimura M. The Plant Organelles Database 2 (PODB2): an updated resource containing movie data of plant organelle dynamics. Plant Cell Physiol. 2011;52:244–253. doi: 10.1093/pcp/pcq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mount DW. Using the basic local alignment search tool (BLAST) CSH Protoc. 2007 doi: 10.1101/pdb.top17. July 1 (doi: 10.1101/pdb.top17; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 24.Camacho C. BLAST+: architecture and applications. BMC Bioinformatics. 2008;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]