Abstract
The PeroxiBase (http://peroxibase.toulouse.inra.fr/) is a specialized database devoted to peroxidases’ families, which are major actors of stress responses. In addition to the increasing number of sequences and the complete modification of the Web interface, new analysis tools and functionalities have been developed since the previous publication in the NAR database issue. Nucleotide sequences and graphical representation of the gene structure can now be included for entries containing genomic cross-references. An expert semi-automatic annotation strategy is being developed to generate new entries from genomic sequences and from EST libraries. Plus, new internal and automatic controls have been included to improve the quality of the entries. To compare gene structure organization among families’ members, two new tools are available, CIWOG to detect common introns and GECA to visualize gene structure overlaid with sequence conservation. The multicriteria search tool was greatly improved to allow simple and combined queries. After such requests or a BLAST search, different analysis processes are suggested, such as multiple alignments with ClustalW or MAFFT, a platform for phylogenetic analysis and GECA’s display in association with a phylogenetic tree. Finally, we updated our family specific profiles implemented in the PeroxiScan tool and made new profiles to consider new sub-families.
INTRODUCTION
Peroxidases are universal enzymes present in all organisms capable to catalyse redox reactions, thanks to various peroxides as electron acceptors. They are encoded by small or large multigenic families and divided into haem or non-haem containing proteins without any evolutionary relationship between the two groups. They are essential for the regulation of reactive oxygen species levels and for the promotion of various substrates oxidation. They play major roles during defence and development processes.
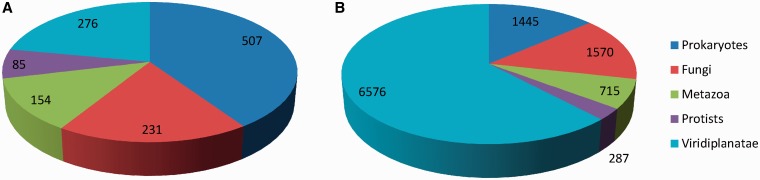
To have a global overview on peroxidases, the PeroxiBase was created in 2004 initially to centralize data concerning class III peroxidases from Viridiplantae. Currently, the numbers of classes, families and superfamilies has largely increased and cover most of the proteins able to reduce peroxides. All kingdoms are well represented in the PeroxiBase (Figure 1A). With the large number of organisms and sequences, the database has become a reference in the field of peroxidases and gene families. It is cross-referenced in UniProt (1) since 2006 and, more recently, in the Arabidopsis database TAIR (2).
Figure 2.
Schematic representation of the workflow for collection, annotation and analysis of sequences. Each step of the individual curration and tools available for the analysis are described elsewhere.
Figure 1.
Numbers of organisms in the PeroxiBase in the five major kingdoms (A) and numbers of entries in the PeroxiBase in every kingdom (B).
Although several databases centralize entries of protein families [CAZy (3), MEROPS (4), ThYme (5) and so forth], the PeroxiBase is unique as being not only a specialized repository of public sequences, but a repository of sequences deduced from expert annotations. Indeed, whole automatic genome annotation generates a number of erroneous sequences, notably with gene merging or splitting problems. This is all the more true when dealing with specific families prone to tandem duplications, such as the peroxidases. Thus, the PeroxiBase is characterized by an expert sequences annotation procedure, with manual curration, which is a guarantee of quality necessary for performing phylogenetic analysis. Efforts to provide only expert annotation derived sequences, in opposition to automated ones exist elsewhere, but only focusing on specific organisms as Vega for Vertebrates (6) or GeneFarm for Arabidopsis (7), which is no longer maintained.
Regarding the peroxidases families, a recent database, PREX (http://csb.wfu.edu/prex/), dedicated to one type of the non-haem peroxidases has been created (8). PREX is merely devoted to subfamily assignment, without re-annotation attempt, but it brings structural and sequence information complementary to those found in our own database.
The PeroxiBase is a dynamic database, constantly being updated with new tools and functions, as well as daily additions of new sequences. To be capable to continue exploring new available sequences and retrieve missing peroxidases, there was an urge for the database to evolve. Thus, the initial interest to centralize high-annotation quality for peroxidases from all living organisms is still our major concern, but the flood of genomic sequences has required the development of tools for semi-automatic annotation.
DESCRIPTION OF TOOLS AND FUNCTIONS
Workflow for semi-automatic annotation
To improve the quality and to speed up the annotation process, two different strategies have been developed to generate new entries. One is based on Scipio, an efficient alignment tool able to deal with splice sites (9) to precisely map a protein onto a genome and produce high quality structural annotation. It generates intron–exon coordinates, DNA and Coding DNA Sequence (CDS) sequences that are directly transferred to the corresponding PeroxiBase entry. The creation of the genomic cross-reference has also been included in this process. A second strategy, based on an approach combining BLAST (10) and CAP3 (11), produces assembled sequences from Expressed Sequence Tag (EST) libraries. This allows the addition of new expression data (numbers of ESTs, tissue types and so forth). This pipeline has been developed with the Galaxy workflow management system (12) and could be easily extracted and used. Both strategies are not visible from the database interface, but are used routinely by the members of the PeroxiBase team to verify and add new sequences and can be shared with all contributors.
Family specific profiles
Expert and specific Hidden Markov model (HMM) -like profiles have already been prepared to define the main peroxidases families and classes (13) and have been implemented in the PeroxiScan tool. But with the increasing number of entries, an update of our family specific profiles had to be done for them to be more representative. Furthermore, new profiles were designed to consider new sub-families. Currently, 80 profiles are installed on the PeroxiBase that cover all kingdoms. These profiles were designed using original data sets obtained by clustering after phylogenetic analysis. Plus, key residues of each subfamily were taken into consideration in these profiles to make them even more specific. Defining these sub-classes and designing the specific corresponding profiles should be of great help in understanding the evolution of this superfamily. It should, in the same time, contribute to a better description of the sequence characteristics in terms of conserved residues, and thus be determinant for an efficient annotation protocol. To promote the use of these profiles, they are also available from MyHits (http://myhits.isb-sib.ch/).
New tools for evolution analysis
One of our goals is to obtain a comprehensive image of the evolution of peroxidases superfamilies that share common ancestral sequences. To try to establish an evolutionary scenario, we provide tools to link heterogeneous data, such as protein and nucleic sequence similarity, gene structure, presence of key residues, localization on the chromosomes and duplication events. For alignment, ClustalW and MAFFT can be used directly online after multicriteria search, and a connection to the Phylogeny website (http://www.phylogeny.fr) allows a phylogenetic analysis with PhyML (14).
In parallel, gene exon/intron organization can be analysed with CIWOG (15) and GECA (16). The latter is a software developed by our team that represents gene exon/intron organization and highlights changes in gene structure among members of a gene family. It is based on protein alignment and is completed with the identification of common introns in the corresponding genes performed by CIWOG. The originality and the relevance of GECA are to overlay a comparison of several gene structures with a symbolic display of their conservation at the sequence level.
As we are convinced that this information can play a major role to elucidate evolutionary history, a pipeline including GECA and PhyML was developed; it generates a phylogenetic tree associated with the gene structure display produced by GECA. The tree can be visualized directly on the PeroxiBase interface thanks to Archaeopteryx (17), a java application for the visualization, analysis and editing of potentially large and highly annotated phylogenetic trees.
New data and check-in for each entry
Thanks to the new annotation procedures developed, nucleotide sequences (genomic, cDNA and CDS) can be found in entries when available. The position on the genomic sequence, and a direct link to the dedicated genome browser, is also provided, plus the gene structure information, in GenBank format, is displayed along with a schematic representation.
New cross-references with a particular attention to ‘Omic’ links have been included to centralize data for comparative genomic analysis. To guaranty the accuracy of the entries, we developed a protocol to verify the submission of new entries, the consistency between the protein, cDNA, CDS, genomic sequences and the intron/exon structures is automatically verified during the submission process. We have also set-up an automatic control of the validity of the cross-reference links during the submission and a semi-annual verification for the existing entries.
New Web interface
The addition of new tools, functions and data in each entry was an instigator to re-design the Web interface. The new version of the PeroxiBase has a modern and easy to use portal. The multicriteria search tool was greatly improved to allow several methods for refining and limiting a query as well as combining several queries. It allows evolutionary analysis thanks to the availability of a package of tools developed by our team and existing ones that are directly usable through the database website.
The PeroxiBase is hosted by the GenoToul bioinformatics facility (http://bioinfo.genopole-toulouse.prd.fr) that allows the use of a computing cluster and makes the database powerful for complex phylogenetic analysis.
DISCUSSION AND FUTURE PROSPECTS
Despite the homogeneity of the organisms distribution within the kingdoms and the increasing number of sequences [from 6026 in 2008 (13) to 10 710 in 2012], the PeroxiBase is still mainly composed of sequences originated from Viridiplantae (62%) (Figure 1B). Although it seems that families of plants peroxidases are more prone to large duplication events, we must continue our effort to balance the representation of families from organisms other than plants. We also need to increase the number of sequences from exotic and poorly represented organisms to perform more global evolution analysis.
The quality of the entries needs to be maintained, but complete manual annotation that guaranties the quality does not allow an efficient coverage of all the sequences available. The semi-automatic protocols developed will facilitate the collection of peroxidase-encoding sequences originated from EST libraries and annotated or non-annotated genomes without lowering the quality of the database. However, the annotation procedure relying on Scipio is only suited for prediction by homology between related or close organisms. To allow prediction on more divergent genomes, we are working on a new strategy that will take advantage of our peroxidases-specific profiles.
Peroxidases families are prone to high tandem, segmental and chromosomal duplication events, which complicates the comprehension of their evolution. The visualization of inter- or intraspecies sequence similarity together with their localization should help to solve this evolution enigma. Choosing between the many available tools for genome browsing and/or comparative genome visualization [see (18) for a review] is our current priority that will lead to the next major change in the PeroxiBase.
FUNDING
Funding for open access charge: LABEX grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank all the past contributors and curators of the PeroxiBase. The PeroxiBase is hosted by the Toulouse Midi-Pyrénées bioinformatics platform. This work has been done in the Plant Science Research Laboratory (LRSV), part of the ‘Laboratoire d’Excellence’ (LABEX) entitled TULIP (ANR-10-LABX-41).
REFERENCES
- 1.Consortium U. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantu DC, Chen Y, Lemons ML, Reilly PJ. ThYme: a database for thioester-active enzymes. Nucleic Acids Res. 2011;39:D342–D346. doi: 10.1093/nar/gkq1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aubourg S, Brunaud V, Bruyère C, Cock M, Cooke R, Cottet A, Couloux A, Déhais P, Deléage G, Duclert A, et al. GeneFarm, structural and functional annotation of Arabidopsis gene and protein families by a network of experts. Nucleic Acids Res. 2005;33:D641–D646. doi: 10.1093/nar/gki115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soito L, Williamson C, Knutson ST, Fetrow JS, Poole LB, Nelson KJ. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2011;39:D332–D337. doi: 10.1093/nar/gkq1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller O, Odronitz F, Stanke M, Kollmar M, Waack S. Scipio: Using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinformatics. 2008;9:278. doi: 10.1186/1471-2105-9-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koua D, Cerutti L, Falquet L, Sigrist CJA, Theiler G, Hulo N, Dunand C. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 2009;37:D261–D266. doi: 10.1093/nar/gkn680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 15.Wilkerson MD, Ru YB, Brendel VP. Common introns within orthologous genes: software and application to plants. Brief. Bioinform. 2009;10:631–644. doi: 10.1093/bib/bbp051. [DOI] [PubMed] [Google Scholar]
- 16.Fawal N, Savelli B, Dunand C, Mathé C. GECA: a fast tool for gene evolution and conservation analysis in eukaryotic protein families. Bioinformatics. 2012;28:1398–1399. doi: 10.1093/bioinformatics/bts153. [DOI] [PubMed] [Google Scholar]
- 17.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen CB, Cantor M, Dubchak I, Gordon D, Wang T. Visualizing genomes: techniques and challenges. Nat. Methods. 2010;7:S5–S15. doi: 10.1038/nmeth.1422. [DOI] [PubMed] [Google Scholar]