Abstract
The cell: an image library-CCDB (CIL-CCDB) (http://www.cellimagelibrary.org) is a searchable database and archive of cellular images. As a repository for microscopy data, it accepts all forms of cell imaging from light and electron microscopy, including multi-dimensional images, Z- and time stacks in a broad variety of raw-data formats, as well as movies and animations. The software design of CIL-CCDB was intentionally designed to allow easy incorporation of new technologies and image formats as they are developed. Currently, CIL-CCDB contains over 9250 images from 358 different species. Images are evaluated for quality and annotated with terms from 14 different ontologies in 16 different fields as well as a basic description and technical details. Since its public launch on 9 August 2010, it has been designed to serve as not only an archive but also an active site for researchers and educators.
INTRODUCTION
After the success of the protein and nucleic acid sequence databases, it was realized that a valuable aspect of researchers’ efforts, microscopic images, needed a similar database. Specialized databases like the Cell Centered Database (1) were created for emerging technologies like electron tomography, but were not equipped to handle individual images from laboratories. There were many extant collections of specific types of images from researchers and there was great interest in having these collections be broader and easily shared. In addition, unpublished image data sequestered on laboratory computers represented a vast but untapped resource that could be of great value to the research and educational communities. There was clearly a need for a resource that could collect large numbers of research images and to share them across a variety of different platforms. Advances in technology rendered these challenges manageable the use of standardized vocabularies from BioOntologies as well as semantic ontologies from the Semantic WEB enables searching with simple terms for the curious as well as more parsed searching for the expert. The recent merger of the cell: an image library (CIL) with the Cell Centered Database from the National Center for Microscopy and Imaging Research at the University of California, San Diego, strengthens the development of CIL-CCDB as a resource and a tool for the cell biology community. This resource is early in its development, as the initial 2 of its 3 years was devoted to setting up the infrastructure and recruiting, vetting and annotating images while incorporating changes based on early user suggestions. While powerful search engines also exist for images and videos, such as Google™ Images and Google™ Videos, CIL-CCDB has significant advantages that increase its effective use for researchers, educators and the general public. These advantages are detailed below. Thus, CIL-CCDB and the general search engines are complementary with CIL-CCDB providing users images and videos that have undergone quality control by cell biology experts as well as detailed information about the images in an easily accessed manner within the database itself.
APPROACH
Although CIL-CCDB’s roots are in research, it was realized early on that the striking nature of cell images provided the opportunity to create a tool that not only was useful for scientific research but also would engage the general public for education and outreach. Developing CIL-CCDB as a robust scientific resource is now coupled with its development for use by educators and the general public.
RESEARCH AND EDUCATION
A number of features were developed to aid users in finding the information they seek or to simply explore CIL-CCDB with its user-friendly interface.
INTERACTIVE CELL AND ORGANELLES
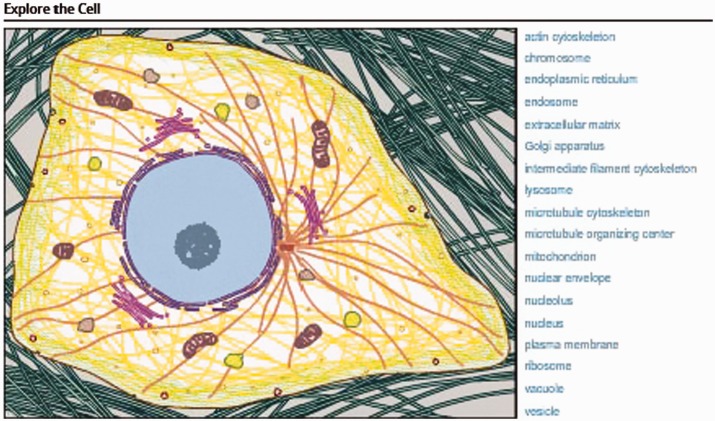
Key to the success of CIL-CCDB is its ability to bridge the abstract concepts of cell biology and cell structure with the reality of that seen in the laboratory. This is most notably demonstrated by the interactive cell illustration on the homepage that invites users to ‘Explore the Cell’ (Figure 1). This feature, unavailable with general search engines such as Google Images, provides easy access to narrative and to images and videos for both non-scientists and experts. By hovering over a cellular feature on the illustration or hovering over the word on the right, users see both the word and all those features highlighted in the illustration. Simply clicking starts an active search for microscopic images of that feature or organelle. Recently added for a number of these features is a second level of interaction. For example, selecting the mitochondrion takes one to a Mitochondrion page with a new interactive illustration of the mitochondrion, a brief explanation of its function, the five most commonly used annotation terms for the Molecular Functions and the five most commonly used annotation terms for the Biological Processes. These annotation terms are from the Gene Ontology (2). The annotation terms in both cases are hyperlinks to searches of all images with that term in CIL-CCDB. Microscopic images of mitochondria then follow this new information. Thus, the user goes from the illustration to the raw data, emphasizing the types of cell images that give rise to the illustrations one sees in textbooks. While there is a difference of opinion about using terms from Gene Ontology, the advantage is that these position CIL-CCDB to create links directly from an image or video into the nucleic acid and protein databases, Genbank and PDB. The intention is to allow users to move from the cell to the molecules and from the cell to the organism via federation with other databases as has been modeled by the Neuroscience Information Framework (NIF) (3). The more general search engines do not provide these types of connections.
Figure 1.
Interactive cell illustration.
BROWSING
In addition to browsing CIL-CCDB with the graphical interface, there is a browse bar located at the top of the page for users to explore CIL-CCDB by different categories, including Cell Process, Cell Component, [(both from the Gene Ontology], Cell Type, [from the Cell Type ontology (4)], Organism [from the NCBI organismal classification ontology (5)] or Recent images. The results pages for this sorting can be customized to sort by a variety of different parameters, including thumbnails of images, alpha order or even the number of images populating a category in the library. These customized sortings are unavailable with the general search engines.
SIMPLE SEARCH
At the top of every page is a simple search box. This search engine is another way to begin accessing the data in CIL-CCDB. After three characters are entered in the search box, suggested terms are present for selection. Only terms that will actually return a result are suggested to prevent the selection of a term with a null result.
ADVANCED SEARCH
The advanced search offers a great deal of power to the user. Images can be searched based on multiple specific attributes such as whether it is a still image, a video or an animation, whether it is 3D with a Z dimension or whether it is a time series. These can be combined for even more selective searching. For example selecting both the Z-stack and the Time Series filters will return those images that have data in ‘both’ the Z dimension of space and the time dimension. These images can be explored further in a multitude of ways. For example, one can move to a certain plane in the Z dimension and watch what happens through time or one can move to a certain time point and move through the different spatial planes.
LICENSING—IMAGES AS DATA
Images, videos and animations are presented in the Library with a variety of licensing options. While the software we have used to build the Library is built on OMERO (6), an open source platform, and our modifications and enhancements are also open source, it was recognized that to be as inclusive as possible it would be best to be able to present images under a wide array of licensing terms. This allows CIL-CCDB to work with not only the classic collections but also publishers, both those that still publish traditionally and those that are beginning to embrace open source publishing. The five licenses used by CIL-CCDB are listed below and as new licensing methods are developed they can easily be incorporated into CIL-CCDB.
LICENSING

Public domain
This image is in the public domain and thus free of any copyright restrictions. However, as is the norm in scientific publishing and as a matter of courtesy, any user should credit the content provider for any public or private use of this image whenever possible. ‘Learn more’.

Attribution only
This image is licensed under a Creative Commons Attribution License. View License Deed | View Legal Code.

Attribution non-commercial share alike
This image is licensed under a Creative Commons Attribution, Non-Commercial Share Alike License. View License Deed | View Legal Code.

Attribution non-commercial; no derivatives
This image is licensed under a Creative Commons Attribution, Non-Commercial, No Derivatives License. View License Deed | View Legal Code.

Copyright
This image is copyright protected. Any public or private use of this image is subject to prevailing copyright laws. Please contact the content provider of this image for permission requests.
Due to the nature of the Creative Commons licensing, the end result of this approach is that we are free to use images from multiple sources, including publishers that are licensed under the Creative Commons licensing, so long as we also present those images with the same licensing.
This approach was also taken to allow us to encourage our image contributors to make their images freely available using the public domain license, while providing them whatever protections they require for their work. It should be noted however, that all images generated by US government employees are automatically in the public domain. However, if one author on a paper is not a federal employee, then the images are protected by whatever licensing applies to the journal in which they are published.
While an author or authors may present a select number of images in their paper, and perhaps even more in the supplemental information, a significantly larger number of images were obtained that led them to their discovery or interpretation. Since these are not published, they are not under any licensing constraints. It is the position of The Cell that entire experimental sets should be submitted, both for archiving and so that others might benefit. Features have been developed so that these, most likely, visually similar images do not overwhelm the library but rather are accessible within a group via an additional click once the representative image for that experiment has been found using the previously described search methods.
Further, CIL-CCDB can present images that have not been publicly available and once published in CIL-CCDB, they become available to the more general search engines while also providing a system to allow the authors of these images to get credit for their publication in CIL-CCDB.
STRUCTURE
A major philosophy of CIL-CCDB is embodied in its approach to presenting information at no cost to the user. Tied to this is also our support of open source software development and open source publishing. While we have incorporated the full range of licensing to ensure that we are able to work with all interested parties, we took a much more open approach in our choice of software to build CIL-CCDB platform. CIL-CCDB, at its core, uses the OMERO platform as its backend image and annotation management system. OMERO is part of the Open Microscopy Environment (7,8) (http://www.openmicroscopy.org/site) suite of products.
The OMERO system has been engineered to work with the Open CCDB tools and infrastructure, maintained by NCMIR at the Center for Research in Biological Systems at UCSD.
There are two methods one can use to contribute images for evaluation for presentation in CIL-CCDB: the DataRollup (DR) and the Online Uploader. The DR is a separate software program that image contributors install on their computers to submit images. It is designed for submitting large numbers of images or for images in the less common formats. It incorporates Bio-Formats (9) (http://loci.wisc.edu/software/bio-formats) and can handle over 100 file formats. The Online Uploader allows users to quickly and easily upload an image directly from the web. Currently, the supported formats include JPG, GIF, TIFF, PNG, PPM, IMOD (.st,.rec,.mrc) and uncompressed AVI. Both methods include the requirement to select the license that should be associated with the image and both allow for the attachment of additional information to aid annotators in completing the entry for that image. The attached documents are never made available to the public so there are no concerns over copyright issues on attached documents. Besides the attached documents we request the image submitter to provide not only their contact information and how the image should be attributed but also to provide some basic information to assist the annotators in generating the entry. This basic information includes information about the biological source (organism, cell type, cellular components, and fixation method and sectioning if applicable), the biological context and probe (e.g. mitotic spindle during division labeled with tubulin primary antibody and Alexa 488 secondary or EGFP-tubulin), the equipment used (e.g. microscope manufacturer, objective with NA, camera, confocal), the acquisition parameters for time series (e.g. taken at two frames per second or confocal Z-stack taken with 1 Airy unit slices), post-processing of still images, if any, and magnification of image (either a scale bar or a note with microns/pixel).
AN API has been developed to assist contributors in automatically submitting images to CIL-CCDB as well as to automatically call data from CIL-CCDB.
Annotation module
Once images are submitted to CIL-CCDB they appear in the annotation module (AM). Annotators complete a number of fields in the AM before publishing the image to the public library. Many of these fields are built on various ontologies. Currently there are 14 different ontologies used in 16 different fields as seen in Table 1. More than just a way to control the vocabularies necessary for meaningful searching, the ontologies, with their inherent relationships between terms, have positioned CIL-CCDB as a collection capable of full participation in the semantic web. As described below, CIL-CCDB is already accessible through portals like the NIF (http://neuinfo.org), which take advantage of these ontological mappings to integrate data across databases.
Table 1.
Ontologies used and the number of fields in which they are used
| Ontology | Number of fields | Web address | References |
|---|---|---|---|
| NCBI organismal classification | 1 | http://bioportal.bioontology.org/ontologies/1132 | PMID: 22139910 |
| Cell type | 1 | http://bioportal.bioontology.org/ontologies/1006 | PMC551541 |
| Cell line ontology | 1 | http://bioportal.bioontology.org/ontologies/1245 | PMID: 18694200 |
| Gene ontology | 3 | http://www.geneontology.org/ | PMID: 22102568 |
| Human development and anatomy | 1 | http://bioportal.bioontology.org/ontologies/1021 | PMID: 22973865 |
| Mouse gross anatomy and development | 1 | http://bioportal.bioontology.org/ontologies/1010 | |
| Xenopus anatomy and development | 1 | http://bioportal.bioontology.org/ontologies/1152 | PMID: 18817563 |
| Zebrafish anatomy and development | 1 | http://bioportal.bioontology.org/ontologies/1051 | |
| Plant growth and developmental stage | 1 | http://bioportal.bioontology.org/ontologies/1587 | PMID: 18194960 |
| Teleost anatomy and development | 1 | http://bioportal.bioontology.org/ontologies/1110 | PMID: 20547776 |
| Human disease | 1 | http://bioportal.bioontology.org/ontologies/1009 | PMC2709267 |
| Infectious disease | 1 | http://bioportal.bioontology.org/ontologies/1092 | Ref (10) |
| Mouse pathology | 1 | http://bioportal.bioontology.org/ontologies/1031 | European mutant mouse pathology database (Pathbase), Pathbase web site, University of Cambridge, World Wide Web (http://www.pathbase.net) October 30, 2012. |
| Biological imaging methods | 8 | http://bioportal.bioontology.org/ontologies/1023 |
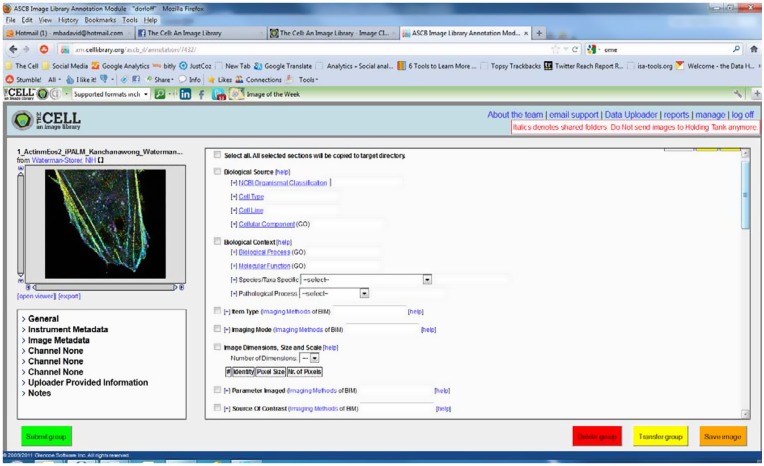
A number of features were built into the AM to increase the ease of using the ontologies for annotation. One key factor of the system is that as an annotator starts typing a term, after the third character, they are presented with matching terms from either that ontology or from that branch of the ontology which makes for easy selection. In addition, all terms have the ability for the annotators to add an additional field so that they can provide more terms for the full description of the image. For example, if the image is described by both Cellular Components ‘lamellipodium’ and ‘actin cytoskeleton’ it is a simple matter for the annotator to add a second field for Cellular Component in the AM and then select these two terms. Figure 2 shows a view of the AM.
Figure 2.
The annotation module.
Development of biological imaging methods ontology
In preparation of developing the AM, a number of different ontologies were researched to determine the most applicable and most widely used. There was not a well-developed ontology describing biological imaging methodologies. We were offered an early version of what was available from Flybase for further extensive development. The Biological Imaging Methods ontology can be found at http://bioportal.bioontology.org/ontologies/1023. This article is the first reference to this ontology.
Publication to CIL-CCDB
Once an image is completely annotated it is published to the public Library, http://www.cellimagelibrary.org.
It was determined that there were a number of circumstances where images in the Library might be better presented to the public if they were grouped. This feature is not available with the more general search engines. Grouping is useful in a number of ways. If images were published in the same article or if images were combined to make a multi panel figure in a publication it would be useful to the user to have a way to see the group. In cases such as this, images are tagged in CIL-CCDB and each one has a line in the entry that says, ‘This image is part of a group’. Selecting the word ‘group’ will now take the user to a page where all the images are presented. Key to this page is also the option to download selected images or all the images in the group. These images can be downloaded in either OME-TIFF file format or as the files that were originally submitted to CIL-CCDB.
As additional grouping scenarios started to arise it was determined that it would be advantageous if there was another grouping option. In some cases, many images would be submitted to the Library that were visually very similar but that were quantitatively different. Even a small group of 200 images would become very unwieldy as a user would browse or search and see pages and pages of what look like very similar or identical images. In response to this and in preparation for the growth of high-throughput imaging, another grouping mechanism was developed. These groups would show only one representative image that would be searchable and browsable. The entry of the representative image would be tagged with, ‘This image is representative of a group of similar images’. Selecting the word group then behaves as before, taking the user to a page where the group images are presented with the option for download.
Advanced search
The advanced search gives users the opportunity to search for images using a great variety of aspects of the image, video or animation.
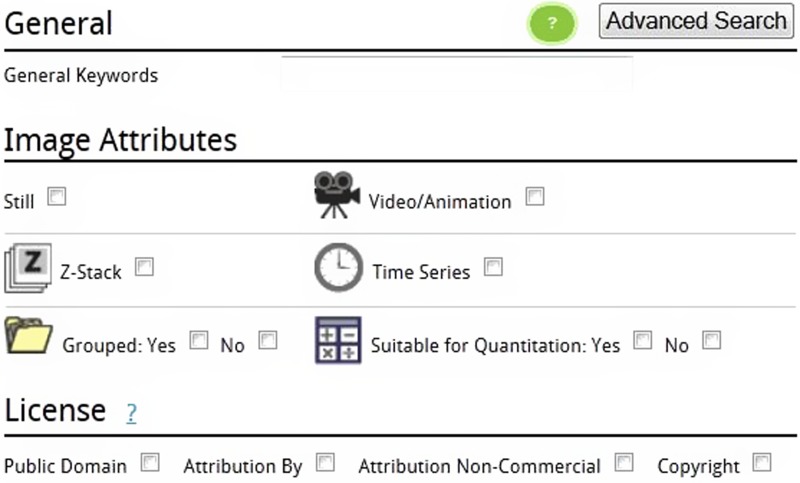
In addition to searching by keyword a user may search by various image attributes. These different attributes can be seen in Figure 3.
Figure 3.
Advanced searching and image attributes.
Note that while there are many images in 3D which includes the Z dimension, some are also stacks of images through time, and still others combine both the Z dimension and the time dimension to create 4D images. These images can also be viewed by holding the time point constant and moving through space in the Z dimension.
The Advanced Search also presents the user with the option to search only for images in groups, a feature which might be useful if, for example, a user is looking for a set of images to use to test a specific pattern recognition or image restoration algorithm (11).
Additionally, the annotators of the images have determined if the images are suitable for quantitation and this information is also a searchable filter. The advanced search also includes a licensing filter. All of these can be combined, so if a user is searching for an image of mitosis for use in a book they could simply find what they are looking for by putting mitosis in the keyword box and selecting both Public Domain and Attribution By. This will return all images that are annotated with mitosis and are either public domain or Creative Commons Attribution only. Since neither of these licenses have any commercial limitations, the user would be free to use the image they select for a variety of commercial or non-commercial purposes (though they are required to note the author of the image as listed in ‘Attribution’ section of the entry if they chose a Creative Commons Attribution only image).
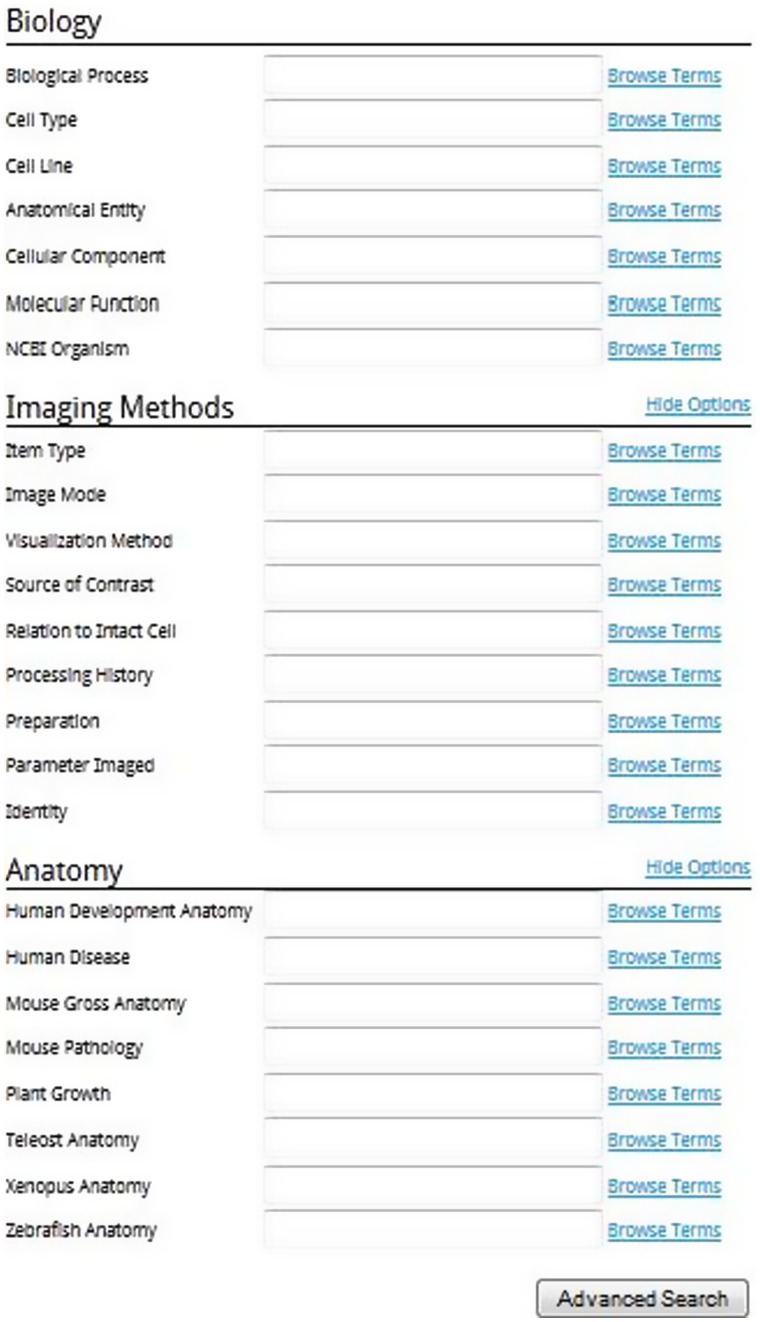
There are also three major scientific headings in the advanced search, Biology, Imaging Methods and Anatomy. These can be seen in Figure 4.
Figure 4.
Scientific headings for the advanced search.
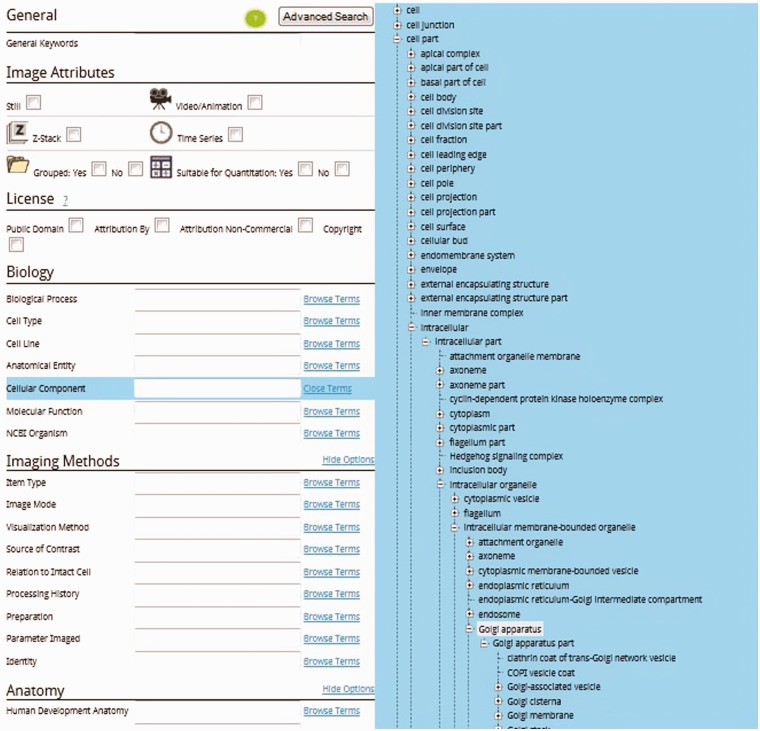
As with the simple search, terms can be typed into these boxes and after three characters the users will be prompted with suggested terms. A key aspect in using the scientific advanced search is the ability to explore the ontology to determine the most appropriate ontology search term to use. The terms presented during the exploration are only those terms that will return one or more images as a result. Figure 5 shows how a user might expand the Cellular Component field by clicking Browse Terms and then expanding the appropriate branches of the ontological tree to reach, e.g. the Golgi apparatus. Simply clicking Golgi apparatus will add it to the advanced search form.
Figure 5.
Advanced search cellular component ontological expansion to Golgi apparatus.
By combining different search terms from the different scientific fields of the advanced search it will be easy to pinpoint those images a user is seeking even as the image library grows to a very large number of images. In the future the user will have the option of subscribing to these searches. That is, once a complex set of search parameters is determined, a user could subscribe to it and receive an email when new images matching those parameters are published in the Library.
ADDITIONAL FEATURES
Open features
A number of other features are available to any user that comes to the site. These features were developed to make it easier for users to move around and find what they want.
Icons in search and browse results
A number of icons are used when presenting search or browse results that give the user information about the image before accessing the detailed image page for that entry. See Figure 6 for the icons and their meanings.
Figure 6.
Icons and their meanings.
Note also that when browsing or searching, the search results also can be filtered by Still, Video/Animation, Z-stack and Time Series. More than one choice may be selected.
Detailed image page
While some features of the detailed image page have already been noted, there are still others. The detailed image page presents the image, video or animation and the licensing. It also has a brief text Description of the image and what is notable. This is followed by the Technical Details that explain how the image was taken or the sample prepared. Often it points to an article that explains the methodologies used. Then the entry lists the various ontological terms annotated in the AM. It should be noted that most of the terms are links that, when clicked, perform a search of the database for all other images that have that same ontological term. There is also a section on the attribution of the image. This should be or is required to be noted when using the image depending on the licensing. This section often contains links to more information and may include a PubMed number linked to an article in PubMed.
Image data download options
On the detailed image page, below the image itself is a button that allows the user to download the image. Images may be downloaded in the JPEG image format, in the OME-TIF image format or the user may download the file that was originally submitted to the Library. For videos and animations the video or animation may be downloaded as a flash video or the file that was originally submitted to the Library. These options give the user great flexibility to use the image for the desired intent.
Web image browser
The Web Image Browser (WIB), developed by the OpenCCDB project (http://openccdb.org) is a tool for interacting with and studying the images in greater detail. As part of a very intuitive interface the WIB gives the user the control over the brightness, contrast and zoom of the image. The WIB was designed to help users navigate very large multispectral images. It also provides simple image processing features such as contrast and brightness adjustment and turning channels on and off. The WIB also has a very simple slide to allow a user to move through the Z dimension or the time dimension when those attributes are present in the data. Users can collaborate over large images by using the collaboration feature by clicking ‘collab’. The WIB also represents the first step the Library has taken to allow its users to annotate regions of interest in the images themselves. This will, in time, enhance our understanding of cell structure and function.
Social media features
There are a number of social media features on each image page, accessed via the social media bar below each image. Using these features, users can email an image with a click of a button or share the image on LinkedIn, StumbleUpon, Facebook, Twitter and numerous other social networks by clicking the Share button. Also available on each page is a Comments section, where users can discuss the images and ask and answer any questions about the images.
Account features
Users of CIL-CCDB are also provided additional functionalities in their accounts. These accounts are free and only require a moment to set up.
In the Profile section of the account is the opportunity to set up Areas of Interest. Again after three characters the user will be given suggestions or these may be free-form text. After adding the Area of Interest and saving the profile changes, images from that and all the defined Areas of Interest will appear on the What’s New page. This effectively gives the account user a customized homepage with a view of what’s new in the Library tailored to specific interests. Images from the last 30 days are presented along with a text link to easily search for earlier images.
Also available to the account user are Photoboxes. These are basically folders created by the user to note flagged images. They can be used in a variety of ways; one might have different photoboxes for different cell processes or perhaps a teacher would have different photoboxes for different lessons.
Interoperability
It has been the approach of the library to distribute the images freely and openly. Partnerships are readily developed between CIL-CCDB and other entities. An API has been developed for external partners to access the images and data of the Library. This feed is designed so that images that the Library presents under the Copyright license are not available to external partners. When these images were submitted to the Library permission was granted for those images to be presented by the Library but the copyright owners still control their use and distribution. The other licenses in use all allow for distribution so long as the license is carried forward. As an open access database, the images, videos and animations of CIL-CCDB also are available to the general search engines that find images and videos. CIL-CCDB.
Partners
Neuroscience information framework
The NIF http://www.neuinfo.org/ is a project supported by the NIH Blueprint Initiative to provide broad access to neuroscience relevant resources (data, tools, materials and services). Through its resource registry and data federation, NIF provides a listing of thousands of resources and federated search across over 170 different data sources. NIF is also built using many of the same core ontologies employed by CIL-CCDB, thus ensuring easy integration of the CIL-CCDB into the NIF data federation. Images from CIL-CCDB are brought together with images from other sources as well. CIL-CCDB also benefits from another of the NIF services, the Linkout service. For those images in the Library that have a PubMed ID, NIF sends data to PubMed that automatically creates a linkout of CIL-CCDB data to that article, using their Link Out feature. These linkouts are found in PubMed by users. So for any images in CIL-CCDB that derive from publications, a link out to these images is provided in PubMed so that users may explore these images further and download them according to the licensing agreements.
DistilBio
DistilBio, (http://distilbio.com/) a life sciences search engine, is another partner that has accessed the data, both the images and the metadata, from CIL-CCDB. They also federate data from a variety of source databases. They have created a search interface to easily query the various databases they have brought together.
IMAGE ACQUISITION
Images have been acquired from a number of sources. Individuals can submit their images directly to the Library using the DR or the Online Uploader. Image sets that informed discovery but that were not published are also sought. In addition to working directly with researchers, we have also acquired images from publishers that support our efforts or those that have licenses that align with ours. As noted earlier, one aspect of Creative Commons licensing is that it allows for sharing and distribution more easily, so long as the licenses are carried forward. Thus CIL-CCDB works with both open access journals as well as some of the traditional publishers that recognize this open access trend and have already begun to adapt their content.
Additionally, we have worked with companies in the fields of cell biology and microscopy since they often have images that are both high quality and stunning. Interestingly, many companies also run image contests for their clients. Some of these are quite prestigious and we are now working to present the winning and honorable mention images from these contests in the Library as well.
CIL-CCDB and google images and videos
The images and videos in CIL-CCDB are also accessed by Google’s search engines and this resource was set up to intentionally have that be the case. This resource was not intended to replace Google Images or Videos. However, the advantages of CIL-CCDB over these Google tools are many. While experts can filter among the images presented by Google, the CIL-CCDB assures that its images have been vetted by professionals enabling experts and non-experts to find material that illustrates cell biology accurately. Images can be analyzed at high resolution within CIL-CCDB, using the WIB available on site and associated with the images. CIL-CCDB’s use of ontologies to categorize and describe images, videos and animations provides intersection with many other databases as well as the semantic WEB facilitating searches beyond that available with the Google tools. There are many images and videos in CIL-CCDB that have not been published and this resource has been designed also as a repository for others to share their work beyond the small numbers of images that may be put into a paper. This aspect also allows researchers to get credit for their work via the citations available in CIL-CCDB. The software that drives CIL-CCDB is open access and available for others to use and adapt. CIL-CCDB has been designed as a research library as well as a library of images for education and the general public; its Groups of images are particularly useful for those working in quantitative and comparative cell biology. There are other advantages of CIL-CCDB for those eager to learn about cell structures and functions from unicellular to multicellular organisms and to see the continuum from cells to molecules and cells to entire organisms. Indeed, the concept of large databases of images and videos of cells complements the databases of nucleic acids and proteins. CIL-CCDB provides an organized, curated and constantly developing and growing resource for database users. Inputs from users hones this resource and increases its utility and such input is encouraged.
CONCLUSION
CIL-CCDB is thus a resource that rides the advancing wave of information and data sharing as well as open access to scientific information funded by public monies. Its growth and development allow not only researchers to develop new insights based on others’ image and video data but also educators and the general public to learn about the complexities and beauties of the biology of cells. User input continues to lead to software adaptations that improve the function and form of CIL-CCDB and much of this feedback comes from the latest in social media. Storage repository, image resource and long-term archiving as technology changes—all these are expectations for CIL-CCDB and the images and videos it makes available. CIL-Cell Centered Database is a work in progress with a substantial number of users from 184 countries. Recent funding assures that this progress continues in developing this resource for the cell biology community and those curious about the workings of cells.
FUNDING
The cell: an image library development supported by the National Institute of General Medical Sciences (NIGMS) [RC2GM092708] of the U.S. National Institutes of Health to the American Society for Cell Biology (to C.M.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS, NIH, NCMIR or ASCB. Open CCDB and WIB development were supported by Open CCDB [GM082949 and RO1NS058296 from the National Institute of Neurological Disorder and Stroke (NINDS) to M.M.] and NCMIR [RR004050 to M.E.]. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all who have contributed in some way to the Library. John Murray (University of Pennsylvania) and Chris Woodcock (University of Massachusetts) developed the Biological Imaging Methods ontology. We thank our Advisory Board for their guidance; we also thank John Murray, for his vision, guidance and dedication to quality, John Hufnagle for his creativity, professionalism and speed at getting things coded, our image archivist, Rachel Scherer of the University of Massachusetts and our annotators for all their hard work and dedication, http://cellimagelibrary.org/pages/personnel. We thank all those who provided their expertise in software development and maintenance for their speed at correcting errors and creating new functionality. We thank those publishers and other partners who took the time to work with us. And, we especially thank those image contributors who took the time to submit their images, work with us to make them available and see and understand the vision of creating this database.
REFERENCES
- 1.Martone ME, Gupta A, Wong M, Qian X, Sosinsky G, Ludascher B, Ellisman MH. A cell-centered database for electron tomographic data. J. Struct. Biol. 2002;138:145–155. doi: 10.1016/s1047-8477(02)00006-0. [DOI] [PubMed] [Google Scholar]
- 2.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imam FT, Larson SD, Bandrowski A, Grethe JS, Gupta A, Martone ME. Development and use of ontologies inside the neuroscience information framework: a practical approach. Front. Genet. 2012;3:111. doi: 10.3389/fgene.2012.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bard J, Rhee ST, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. Published online 14 January 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allan C, Burel J-M, Moore J, Blackburn C, Linkert M, Loynton S, MacDonald D, Moore WJ, Neves C, Patterson A, et al. OMERO: flexible, model-driven data management for experimental biology. Nat Methods. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston J, Nagaraja A, Hochheiser H, Goldberg IG. A flexible framework for web interfaces to image databases: supporting user-defined ontologies and links to external databases. 2006 IEEE International Symposium on Biomedical Imaging. 2006 [Google Scholar]
- 8.Goldberg I, Allan C, Burel J-M, Creager D, Falconi A, Hochheiser H, Johnston J, Mellen J, Sorger PK, Swedlow JR. The open microscopy environment (OME) data model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linkert M, Rueden CT, Allan C, Burel J-M, Moore W, Patterson A, Loranger B, Moore J, Neves C, MacDonald D, et al. Metadata matters: access to image data in the real world. J. Cell Biol. 2010;189:5777–5782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowell LG, Smith B. 2010. Infectious Disease Ontology. Infectious Disease Informatics, Chapter 19, Sintchenko V; pp. 373–395. [Google Scholar]
- 11.Lefkimmiatis S, Bourquard A, Unser M. Hessian-based norm regularization for image restoration with biomedical applications. IEEE Trans. Image Process Source. 2012;21:983–995. doi: 10.1109/TIP.2011.2168232. [DOI] [PubMed] [Google Scholar]