Abstract
A wide range of resources on gene expression profiling enhance various strategies in plant molecular biology particularly in characterization of gene function. We have updated our gene expression profile database, RiceXPro (http://ricexpro.dna.affrc.go.jp/), to provide more comprehensive information on the transcriptome of rice encompassing the entire growth cycle and various experimental conditions. The gene expression profiles are currently grouped into three categories, namely, ‘field/development’ with 572 data corresponding to 12 data sets, ‘plant hormone’ with 143 data corresponding to 13 data sets and ‘cell- and tissue-type’ comprising of 38 microarray data. In addition to the interface for retrieving expression information of a gene/genes in each data set, we have incorporated an interface for a global approach in searching an overall view of the gene expression profiles from multiple data sets within each category. Furthermore, we have also added a BLAST search function that enables users to explore expression profile of a gene/genes with similarity to a query sequence. Therefore, the updated version of RiceXPro can be used more efficiently to survey the gene expression signature of rice in sufficient depth and may also provide clues on gene function of other cereal crops.
INTRODUCTION
Elucidating the function of all predicted genes in the rice genome is an ultimate goal not only for basic research in plant molecular biology but also for applied aspects of genomics to accelerate the improvement of rice that serves as staple food for almost half of the world’s population. The major achievements in elucidating a high-quality genome sequence (1) have been complemented with worldwide initiatives to develop various infrastructures for characterizing the function of rice genes (2–10). However, despite these extensive efforts, more than half of ∼32 000 predicted genes in the rice genome are still classified either as hypothetical or unknown genes because no function has been assigned to their protein products. So far, only ∼600 genes had been fully characterized by map-based cloning and other strategies (11,12).
Information on the specificity of gene expression in terms of organ/tissue and developmental stage that can be obtained by comprehensive gene expression profiling may lead to a thorough understanding of gene function. In rice, large-scale gene expression profiling covering different organs, tissues and cell types at various developmental stages have been reported (13–15). The rapid accumulation of rice microarray data in public repositories such as NCBI’s Gene Expression Omnibus (GEO) (16,17), EBI Array Express (18) and DDBJ CIBEX (19) also led to the development of informatics resources and analysis tools for integrating and extracting useful information on gene expression (20–23). The Rice Oligonucleotide Array Database (ROAD) contains 1867 microarray data available in the public domain and provides functional analysis tools (20). The OryzaExpress features gene expression information integrated with omics data from public databases (21). The rice component of the PlantArrayNet is basically designed for retrieving gene co-expression information in rice (22).
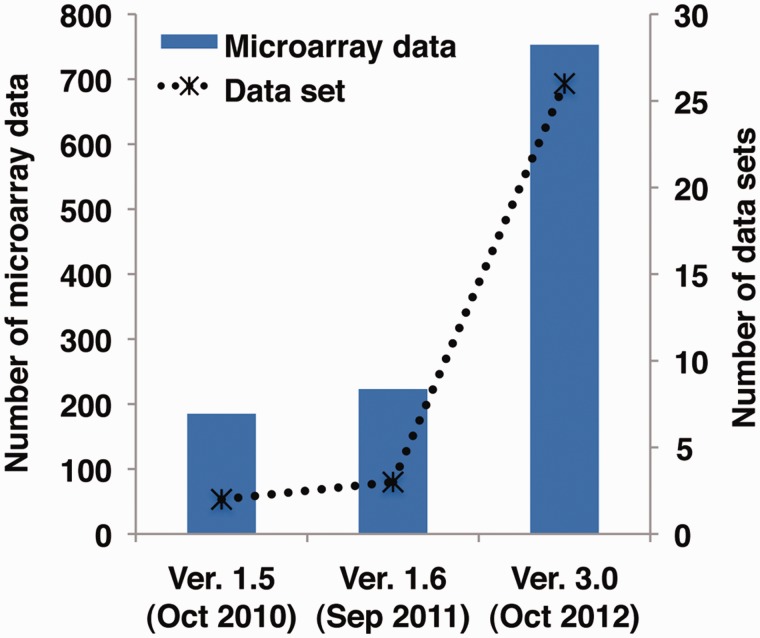
Our database, RiceXPro, was initially conceptualized with the aim of providing a platform for monitoring gene expression of the rice plant under the natural field conditions (24). The first release (version 1.5) originally contained two data sets in the field/development category corresponding to spatio-temporal gene expression profiling based on 48 different tissues and organs at various developmental stages (RXP_0001) and continuous profiling of mature leaves from transplanting until harvesting (RXP_0003) (Table 1, Figure 1). As a minor update (version 1.6), we added a data set on root gene expression profile (RXP_4001) obtained via a combined laser microdissection and microarray approaches (25) and created a data category for ‘cell- and tissue-type’ to distinguish from the two data sets, which were derived from samples grown in the field. The previous versions of RiceXPro included user-friendly interfaces for searching and viewing gene expression profiles, analysis tools and construction of heat maps.
Table 1.
Microarray data in RiceXPro Version 3.0
| Category | Data set ID | Microarray analysis | No. of data | Description |
|---|---|---|---|---|
| Field/development | RXP_0001 | One color | 143 | Spatio-temporal gene expression of various tissues/organs throughout entire growth in the field |
| RXP_0002 | One color | 200 | Diurnal and circadian gene expression profile of leaf throughout entire growth | |
| RXP_0003 | One color | 51 | Leaf gene expression profile throughout entire growth in the field (12:00) | |
| RXP_0004 | One color | 34 | Leaf gene expression profile throughout entire growth in the field (24:00) | |
| RXP_0005 | One color | 28 | Leaf gene expression profile during sunrise | |
| RXP_0006 | One color | 19 | Leaf gene expression profile during sunset | |
| RXP_0007 | One color | 14 | Root gene expression profile throughout entire growth in the field (12:00) | |
| RXP_0008 | One color | 14 | Root gene expression profile throughout entire growth in the field (24:00) | |
| RXP_0009 | One color | 50 | Diurnal and circadian gene expression profile of root | |
| RXP_0010 | One color | 54 | Gene expression profile during reproductive organ development | |
| RXP_0011 | One color | 34 | Grain gene expression profile at early developmental stage | |
| RXP_0012 | One color | 36 | Embryo and endosperm gene expression profile at ripening stage | |
| Plant hormone | RXP_1000 | Two color | 138 | Global gene expression profile in response to plant hormone |
| RXP_1001 | Two color | 18 | Root gene expression profile in response to abscisic acid | |
| RXP_1002 | Two color | 18 | Root gene expression profile in response to gibberellin | |
| RXP_1003 | Two color | 18 | Root gene expression profile in response to auxin | |
| RXP_1004 | Two color | 18 | Root gene expression profile in response to brassinosteroid | |
| RXP_1005 | Two color | 18 | Root gene expression profile in response to cytokinin | |
| RXP_1006 | Two color | 10 | Shoot gene expression profile in response to abscisic acid | |
| RXP_1007 | Two color | 10 | Shoot gene expression profile in response to gibberellin | |
| RXP_1008 | Two color | 10 | Shoot gene expression profile in response to auxin | |
| RXP_1009 | Two color | 10 | Shoot gene expression profile in response to brassinosteroid | |
| RXP_1010 | Two color | 10 | Shoot gene expression profile in response to cytokinin | |
| RXP_1011 | Two color | 18 | Root gene expression profile in response to jasmonic acid | |
| RXP_1012 | Two color | 10 | Shoot gene expression profile in response to jasmonic acid | |
| Cell and tissue type | RXP_4001 | One color | 38 | Root gene expression profile covering various developmental stages and tissue types |
Figure 1.
Microarray data and data sets in previous and current versions of RiceXPro.
Here, we describe an updated version of RiceXPro (version 3.0) with the expansion of data on field transcriptome, addition of microarray data on plant hormone treatment and categorical grouping of data sets for more efficient browsing of expression data. Major improvements in search options include a global search approach to get an overall view of expression profile in each data category, search for expression profile within a specific region of the chromosome and a Basic Local Alignment Search Tool (BLAST)-based gene expression similarity search that can be useful for other cereal crops.
UPDATED DATABASE CONTENTS
We have added a total of 530 microarray data representing 23 data sets (Figure 1). All data have been deposited in NCBI’s GEO (16,17), and accessible through GEO series accession numbers GSE36040, GSE36042, GSE36043, GSE36044, GSE39423, GSE39424, GSE39425, GSE39426, GSE39427, GSE39429 and GSE39432. The RiceXPro Version 3.0 currently contains a total of 753 microarray data representing 26 data sets grouped into three categories, namely, ‘field/development’, ‘plant hormone’ and ‘cell- and tissue- type’. Table 1 provides a list of data sets in each category. Other features of each data set such as search options, data type, graph format, analysis tools and download options are summarized in Supplementary Table S1. All data were generated using a single microarray platform (Rice 4x44K RAP-DB microarray; Agilent Technology).
Field/development category
The first version of RiceXPro consisted of data on spatio-temporal gene expression profiles of tissues/organs (RXP_0001) and leaf expression profiles throughout the entire growth (RXP_0003). With the 10 new data sets, the ‘field/development’ category currently contains a wide range of gene expression data encompassing the entire growth cycle under natural field conditions and more detailed time course data at various stages of development. All data were obtained during the 2008 cultivation season. Some microarray data are included in two or more data sets to provide sufficient interpretation of expression data. For example, 44 microarray data in RXP_0001 are also included in RXP_0010. Although RXP_0001 basically provides information on organ and tissue specificity in gene expression throughout entire growth, RXP_0010 focuses mainly on reproductive organ development, thereby allowing users to select a data set representing appropriate biological process of interest. RXP_0002 is the largest data set with a total of 200 microarray data corresponding to the uppermost fully expanded leaves in a 48-h period at 2-h intervals from eight different growth stages. Therefore, this data set could be useful for elucidating not only the growth stage specificity but also the diurnal rhythmic pattern of gene expression throughout the entire growth in the field. RXP_0004 represents gene expression profile of the leaf at night-time (12 midnight) based on samples collected at weekly interval from 14 to 126 days after transplanting (DAT). We have also incorporated data sets to characterize the gene expression in response to gradual changes of light intensity in the field. These data sets represent leaf samples collected at 10-min interval during sunrise (RXP_0005) and sunset (RXP_0006). We have three data sets focusing on gene expression of root throughout entire growth at daytime (RXP_0007), night-time (RXP_0008) and diurnal rhythm at 2-h interval (RXP_0009). The two data sets focusing on seed development (RXP_0011 and RXP_0012) could be useful in elucidating the molecular mechanisms during the seed development. Taken together, the gene expression data in the ‘field/development’ category of RiceXPro provide baseline information underlying various biological processes throughout entire growth of rice under natural field conditions.
Plant hormone category
In addition to the gene expression data of rice under natural field conditions, we performed microarray analysis of rice plants treated with different plant hormones. The newly added data in RiceXPro include 12 data sets (RXP_1001∼RXP_1012) representing gene expression profiles of shoot and root in response to six plant hormones, namely, abscisic acid, gibberellin, auxin, brassinosteroid, cytokinin and jasmonic acid (Table 1). These data were collected from 7-day-old seedlings and included root samples collected at 15 min, 30 min, 1 h, 3 h and 6 h of incubation and shoot samples collected at 1, 3, 6 and 12 h of incubation after hormone treatment. Root and shoot samples from untreated seedlings were used as control (mock treatment). The gene expression data were generated using a two-color microarray system with the treated samples labeled with Cy5 and the untreated control labeled with Cy3. The time course expression profile for each gene is shown as the log-ratio of signal intensity (Log2 Cy5/Cy3) (line graph) and Cy3 and Cy5 signal intensity (bar graph). To provide a general view of the expression signature in response to various plant hormones, all microarray data for plant hormone treatment (total of 138 data) were integrated into a single data set (RXP_1000). To our knowledge, although a number of genes related to plant hormone metabolism and signaling control agronomically important traits that determine crop yield (26–30), comprehensive gene expression profiles in response to plant hormones had not been available in rice. Therefore, our gene expression profiles on the response to various plant hormones can provide further insights in understanding gene expression signature in combination with profiles in the field/development category.
GLOBAL VIEW OF GENE EXPRESSION PROFILE
In addition to the gene expression search that can be initiated from the interface for each data set, we incorporated a ‘Global Profile’ search option to provide an overall view of the gene expression in each data category. The users can now access gene expression data from two entry points in the top page indicated by respective panels for ‘Data Sets’ and ‘Global Profile’. The data set approach facilitates in-depth retrieval of the expression information of a gene/genes within a data set using keyword search, chromosome search, analysis tools and/or heat map option as previously reported (24). However, this approach requires the user to browse each data set for viewing the gene expression under different conditions. On the other hand, the global approach provides the users with a quick overall view of all the gene expression information among all the data sets within a category as summarized in a single web page.
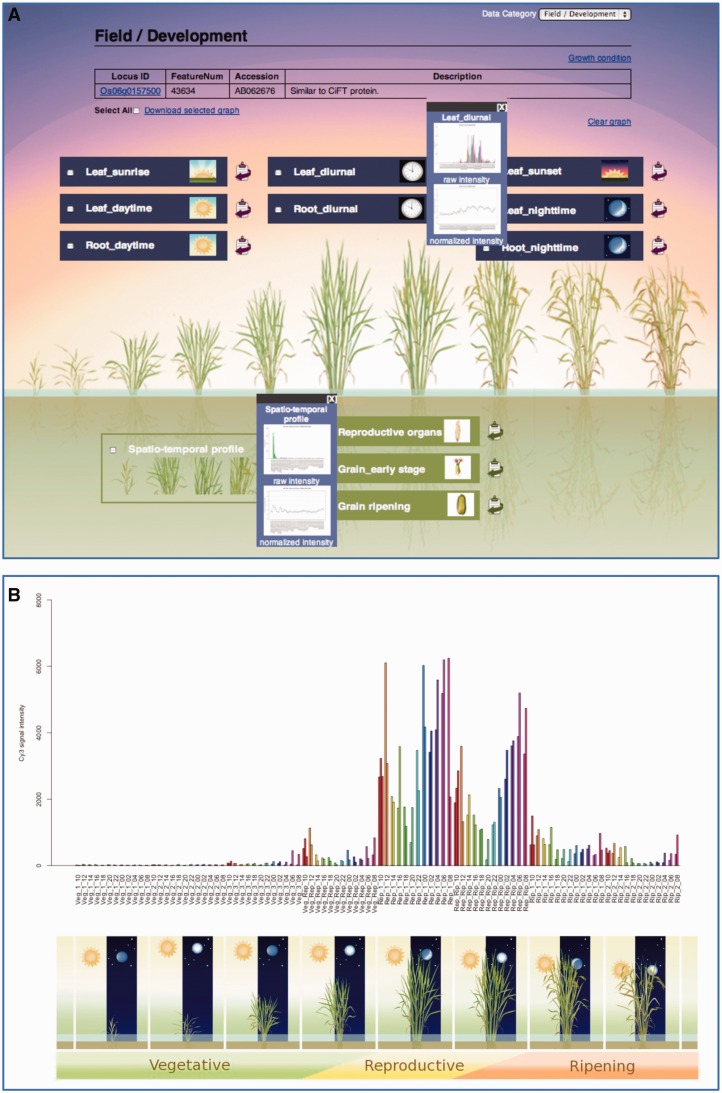
From the ‘Global Profile’ link, users can currently view the overall gene expression profiles from the ‘field/development’ and ‘plant hormone’ categories. The keyword and chromosome search options provide a tabular list of the gene/genes with RAP-DB locus ID (RAP ID), feature number representing the ID number of the probes in the Agilent microarray platform, accession number, probe sequence identifier, gene description and MSU Osa1 Rice Loci (MSU-ID). The feature number provides the link to the interface with image icons for identification of data sets in each category (Figure 2A). With mouseover on an image icon, a pop-up window opens with an overview of the expression profile for each gene in a data set. The ‘sample image’ icon and accompanying ‘data image’ icon also serve as links to the gene expression profile in graph format and the search page of each data set, respectively (Figure 2B). The interface for global profile of the gene provides link to other databases, namely, Rice TOGO Browser (31), RAP-DB (5) and SALAD database (32), via the locus ID, download option for selected graph(s) and switching option to other data category.
Figure 2.
Global profile approach for access to expression data of a gene in multiple data sets. (A) Interface for field/development category in global profile approach. The users can view overall expression profile of a gene within the category from the pop-up windows. The expression profiles of the rice florigen gene, RFT1 (Os06g0157500), in two data sets (RXP_0001 and RXP_0002) are shown in the pop-up windows. Graphical data of gene expression profile is accessible by clicking the image icon. (B) The bar graph representing raw signal intensity of RFT1 in a time course sampling at 2-h intervals for 48 h at eight different growth stages (RXP_0002).
As an example, the global expression profile of RFT1 (Os06g0157500), a rice ortholog of Arabidopsis florigen gene FLOWERING LOCUS T (FT), in the field/development category is shown in Figure 2A. The RFT1 protein synthesized in the leaf blade is transported to the shoot apical meristem and functions as a florigen in long-day condition (33). Based on the spatio-temporal profile (RXP_0001), RFT1 is expressed specifically in the leaf blade at the reproductive stage. The leaf diurnal profile (RXP_0002) provides further insight into the expression signature of this florigen gene (Figure 2B). In particular, this data set shows that RFT1 was induced just before reproductive transition, then increased during the reproductive stage and eventually decreased during the ripening stage. The peak of expression was recorded at dawn (around 6:00 AM). Therefore, the global profile approach would help researches to efficiently explore gene expression profile in a wide range of conditions.
IMPROVEMENT OF SEARCH OPTIONS AND VIEWER
We have added several features in the updated version of RiceXPro to further improve the search functions and gene expression viewers. Furthermore, we have incorporated a homology search option that will allow researchers of other cereal crops to explore the gene expression data in our database.
Region search
The information of position and time specificity on gene expression would enhance map-based cloning in forward genetics strategy. Therefore, in addition to chromosome search, we incorporated search option that would allow the user to characterize gene expression of all the genes in a specific region of the genome. This region search function is initiated by entering the two loci flanking a desired region of the chromosome. A list of genes within the region is generated with links to the expression profiles and other information. This function is available in the search options for both global profile and individual data sets.
Gene expression profile viewer
In any interface for expression profile in graph format, the users can view the graphs in either a scroll mode or a tab mode. Although all signal intensity data can be accessed from the GEO link, we have also provided a link for viewing the raw and normalized signal intensity via ‘View plot data’ link in any page providing gene expression profile in graph format.
BLAST-based expression search
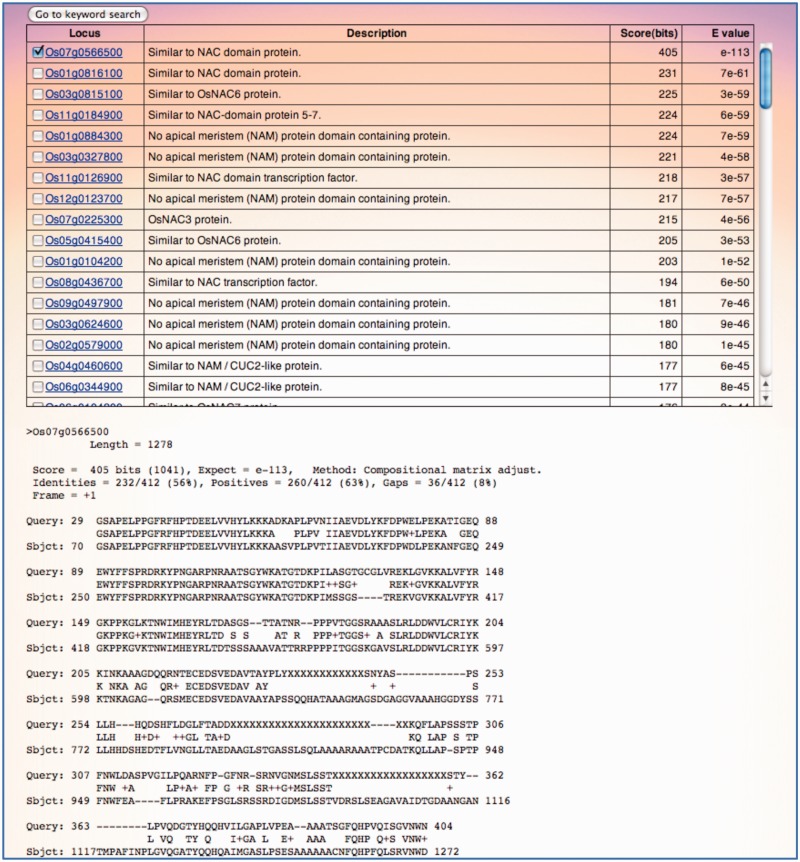
The comprehensive gene expression profile of rice could be useful for researchers of other cereal crops such as wheat, barley and maize. We have incorporated a sequence similarity search, which we named EXP_BLAST, to explore expression profile of a gene/genes with similarity to a query sequence. This search function allows users to extract a gene/genes by BLAST search options (34,35), namely, blastn, tblastn and tblastx against coding sequence of genes with probes in the Rice 4x44K Microarray platform (Figure 3). The search result includes a list of a gene/genes with similarity to the query sequence with the locus ID, description, score, and e-value. Details of the search result for each gene, including alignment information, can be viewed from locus ID. The users can access expression profile of selected gene(s) via global profile approach. The example shown in Figure 3 represents the search result generated using the amino acid alignment of wheat NAM-B1, a QTL gene involved in accelerating senescence and increasing nutrient remobilization from leaves to developing grains (36), as a query sequence. A more detailed survey of the expression data in RXP_0002 and RXP_0003 confirms that the expression of Os07g0566500, the top hit gene in EXP_BLAST search, is observed specifically in leaves during senescence.
Figure 3.
Interface for EXP_BLAST search using amino acid alignment of wheat NAM-B1 as a query sequence. The results page consists of a list of gene/genes with sequence similarity to the query sequence based on score and e-values and the pairwise sequence alignment. The expression profile of the gene/genes can be accessed via keyword search from the global profile search option.
CONCLUSION
The technical protocols for microarray technology have become more standardized and reproducible. This allowed us to perform extensive transcriptome analyses on various aspects of rice growth and development and incorporate the high-quality gene expression information in RiceXPro. Furthermore, as we collected all data in our laboratory using a single microarray system, the gene expression data would be more uniform in all experiments. This provides an opportunity to compare gene expression data across different tissues/organs, different growth stages and different experimental conditions with high reliability.
Growth and development of the rice plant in the field, a much more complex environment than laboratory conditions, is accompanied with various changes in physiological and morphological state of the plant triggered by internal or external stimuli. The transcriptome could be used as an index for monitoring the changes in many biological processes. It is therefore worth understanding the global configuration and complexity of the transcriptome that underlies physiological changes throughout the entire growth in the field to develop strategies for crop improvement. As a major update, we have incorporated a large collection of gene expression profiles of the rice plant from transplanting until harvesting. Therefore, the updated RiceXPro could provide more valuable insights into gene expression networks involved in many biological events throughout growth under natural field conditions that may eventually provide clues for understanding many agronomic traits in rice.
We are currently undertaking microarray analysis involving rice plants subjected to biotic and abiotic stresses, as well as cell and tissue types at various stages of development. These data will be added to the database and functionalities will be enhanced further to provide a more extensive platform for exploring the transcriptome of rice and to expand the utility of RiceXPro as a major source of gene expression information for researchers of rice and other cereal crops.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
Ministry of Agriculture, Forestry and Fisheries (MAFF) of Japan (to Y.N.) through the Genomics for Agricultural Innovation [RTR0002]. Funding for open access charge: RTR0002.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Drs Hidemasa Imazeki, Hiroh Shibaoka, Makoto Matsuoka, Moritoshi Iino, Tsukaho Hattori, Hitoshi Mori and Hitoshi Sakakibara for useful suggestions on plant hormone treatment. We also thank Ms Ritsuko Motoyama for microarray analysis.
REFERENCES
- 1.The International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 2.Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, et al. The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice Annotation Project. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice Annotation Project. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An G, Lee S, Kim SH, Kim SR. Molecular genetics using T-DNA in rice. Plant Cell Physiol. 2005;46:14–22. doi: 10.1093/pcp/pci502. [DOI] [PubMed] [Google Scholar]
- 8.Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. Generation of a flanking sequence tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 9.Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 2007;63:351–364. doi: 10.1007/s11103-006-9093-z. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, Pang J, Higashi N, Ando S, Toki S, et al. A genome-wide gain-of-function analysis of rice genes using the FOX-hunting system. Plant Mol. Biol. 2007;65:357–371. doi: 10.1007/s11103-007-9243-y. [DOI] [PubMed] [Google Scholar]
- 11.Jung KH, An G, Ronald PC. Towards a better bowl of rice: assigning function to tens of thousands of rice genes. Nat. Rev. Genet. 2008;9:91–101. doi: 10.1038/nrg2286. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Cai Z, Xie W, Long T, Yu H, Zhang Q. Rice functional genomics research: progress and implications for crop genetic improvement. Biotechnol. Adv. 2012;30:1059–1070. doi: 10.1016/j.biotechadv.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat. Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61:752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- 15.Sato Y, Antonio BA, Namiki N, Motoyama R, Sugimoto K, Takehisa H, Minami H, Kamatsuki K, Kusaba M, Hirochika H, et al. Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol. 2011;11:10. doi: 10.1186/1471-2229-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeo K, Ishi-I J, Tamura T, Gojobori T, Tateno Y. CIBEX: center for information biology gene expression database. C. R. Biol. 2003;326:1079–1082. doi: 10.1016/j.crvi.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 20.Cao P, Jong KH, Choi D, Hwang D, Zhu J, Ronald PC. The rice oligonucleotide array database: an atlas of rice gene expression. Rice. 2012;5:17. doi: 10.1186/1939-8433-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamada K, Hongo K, Suwabe K, Shimizu A, Nagayama T, Abe R, Kikuchi S, Yamamoto N, Fujii T, Yokoyama K, et al. OryzaExpress: an integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 2011;52:220–229. doi: 10.1093/pcp/pcq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee TH, Kim YK, Pham TT, Song SI, Kim JK, Kang KY, An G, Jung KH, Galbraith DW, Kim M, et al. RiceArrayNet: a database for correlating gene expression from transcriptome profiling, and its application to the analysis of coexpressed genes in rice. Plant Physiol. 2009;151:16–33. doi: 10.1104/pp.109.139030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2012;40:D1194–D1201. doi: 10.1093/nar/gkr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato Y, Antonio B, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39:D1141–D1148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, Minami H, Namiki N, Inukai Y, Nakazono M, et al. Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J. 2012;69:126–140. doi: 10.1111/j.1365-313X.2011.04777.x. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 27.Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 28.Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 29.Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51:1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- 30.Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- 31.Nagamura Y, Antonio BA, Sato Y, Miyao A, Namiki N, Yonemaru J, Minami H, Kamatsuki K, Shimura K, Shimizu Y, et al. Rice TOGO Browser: a platform to retrieve integrated information on rice functional and applied genomics. Plant Cell Physiol. 2011;52:230–237. doi: 10.1093/pcp/pcq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mihara M, Itoh T, Izawa T. SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 2010;38:D835–D842. doi: 10.1093/nar/gkp831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]