Abstract
As an alternative to modern western medicine, Traditional Chinese Medicine (TCM) is receiving increasingly attention worldwide. Great efforts have been paid to TCM’s modernization, which tries to bridge the gap between TCM and modern western medicine. As TCM and modern western medicine share a common aspect at molecular level that the compound(s) perturb human’s dysfunction network and restore human normal physiological condition, the relationship between compounds (in herb, refer to ingredients) and their targets (proteins) should be the key factor to connect TCM and modern medicine. Accordingly, we construct this Traditional Chinese Medicine Integrated Database (TCMID, http://www.megabionet.org/tcmid/), which records TCM-related information collected from different resources and through text-mining method. To enlarge the scope of the TCMID, the data have been linked to common drug and disease databases, including Drugbank, OMIM and PubChem. Currently, our TCMID contains ∼47 000 prescriptions, 8159 herbs, 25 210 compounds, 6828 drugs, 3791 diseases and 17 521 related targets, which is the largest data set for related field. Our web-based software displays a network for integrative relationships between herbs and their treated diseases, the active ingredients and their targets, which will facilitate the study of combination therapy and understanding of the underlying mechanisms for TCM at molecular level.
INTRODUCTION
Traditional Chinese Medicine (TCM) has been widely used in treating various diseases in Eastern Asia for several thousand years. Currently, TCM still plays a critical role in maintaining the health of the Chinese people, and it gains more and more attention around the world. Modern medical field also adopts the ideas of TCM by using combinational drugs to treat complex diseases, such as cancers, diabetes, etc. However, TCM is based on Yinyangism (i.e. the combination of Five Phases theory with Yin-yang theory), which is different from the philosophy of modern western medicine, this largely prevents TCM from being recognized by the western countries. Thus, to facilitate TCM to serve people better, it is essential to bring the ancient practice of TCM into line with modern standards (i.e. understanding the progression and treatment of disease at the molecular level) (1,2).
In recent decades, Hong Kong, Taiwan and mainland China have made great efforts to study various TCMs and decompose their components, from which quite a lot bioactive ingredients have been isolated and identified (3). This has successfully resulted in the discovery of a variety of single compound-based therapeutics, such as artemisinin for malaria treatment and salvicine for anticancer (4). On one hand, these research data are precious resources and can provide important guides for further systematic study. On the other hand, TCM practice takes a more holistic method; the benefits of TCM drugs often come as a result of synergistic interactions of multiple ingredients (5). Therefore, to truly modernize Chinese medicine, systematic methods should be adopted, and all the active ingredients of herbs or formula should be taken into consideration simultaneously. Thus, linking TCM to their targets and treated diseases, which are deeply studied by modern life science, definitely provides useful information and can help us to demystify the theory underlying TCM.
Currently, the mainstream pharmaceutical industry encounters a predicament that the investment has increased substantially in recent years, but the annual number of truly innovative new drugs approved by the US Food and Drug Administration has not increased accordingly (6). An important factor contributing to this dilemma is the robustness of phenotype determined by the redundant functions of related proteins and the alternative compensatory signalling routes (7). The main strategy in drug discovery is based on ‘one gene–one drug–one disease’ paradigm, which is to screen potential compounds for individual disease-causing target; however, the drug’s efficacy is impaired by the robustness of the protein interaction network in the treated objectives. To overcome the limitations, the concept of effective combinatorial drugs and drugs with multiple targets has led to an increased interest in systems-oriented approaches for drug discovery (8). Considering the difficulties to develop a single compound into a medicine, the development of a combination of compounds will be exponentially more complex. Thus, turning to TCM for inspiration will be a sensible solution, as TCM treats diseases or dysfunctions in a more holistic way. The main therapeutic approach of TCM is to use herbs or formula (mixture of herbs) that contains hundreds, even thousands of compounds, to make the organism rebalanced. Therefore, at the molecular level, TCM formulae are multi-component and multi-target agents, which is essentially the same strategy as the combination therapy of multi-component drugs.
Due to the reasons mentioned above, we built this TCM-integrated database to host data on all aspects of TCM and linked them to the related results of modern western medicine. As TCM and modern western medicine share a common goal of maintaining human health, and they all treat diseases by using compounds to interact with disease-specific function networks, we use this common aspect as a key factor to bridge the gap between TCM and modern western medicine.
Although there are many databases recoding information for TCM sources, yet these databases either lack connections between herb ingredients and targets or the number of records in those databases is not comprehensive enough for systematic analysis. TCM-ID (9) consists of only 1588 prescription, 1313 herbs and <6000 ingredients, and there is no information about the connections between ingredients (compounds) and targets. Although HIT (10) comprises information about these connections, the number of herbal ingredients it records is only 586 with 1301 related targets. TCM@Taiwan (11) contains detailed information on a large number of herbal ingredients, but the knowledge on related herbs and targets is not collected. Other databases, such as TCMD (12), TCMGeneDIT (13) and Chinese Traditional Medicine Herbs Database (14), are either not well organized or not free for sharing information. Through comprehensively integrating various data and information, our Traditional Chinese Medicine Integrated Database (TCMID) stores records about >8000 herbs, 25 000 herbal compounds, 17 500 targets and other related information for TCM as well as modern medicine (Table 1). To our knowledge, our TCMID is the largest database for the TCM field with a great improvement on information integration. In addition, we developed a network display tool to virtually present their connection to facilitate the understanding of mechanism underlying TCM and the research of combination therapy.
Table 1.
Data resources
| Data field | Date source | Amount of data |
|---|---|---|
| Prescription | Text-mining | 46 914 |
| Herb | TCM-ID, text-mining | 8159 |
| Ingredients | TCM-ID, HIT, TCM@Taiwan, text-mining | 25 210 |
| Targets | HIT, STITCH, OMIM, DrugBank, text-mining | 17 521 |
| Disease | OMIM | 3791 |
| Drugs | DrugBank | 6826 |
MATERIALS AND METHODS
TCMID is composed of six data fields, namely prescriptions, herbs, ingredients, targets, drugs and diseases. The information and data in those fields were integrated from related web-based databases and text mining of books and published articles.
The prescriptions were collected mainly through text mining from books and published articles. Information for herbs was mainly extracted from TCM-ID database and referred to a book—Encyclopedia of Traditional Chinese Medicines (15). The data field about herbal ingredients, such as name and structure, was inputted by combining information from TCM@Taiwan, TCM-ID and Encyclopedia of Traditional Chinese Medicines. Information of diseases and their related proteins, drugs and their targets was retrieved from DrugBank and OMIM. As the target ID used by DrugBank, OMIM and other sources are different from each other, the data from those resources are inconsistent and incomparable. To overcome the barriers, we converted all of them into UniProt AC—a comprehensive, high-quality and freely accessible resource of protein sequence and functional information (16).
The main goal of our system is to build the connections between the herbal ingredients and diseases through disease genes/proteins, which could also be potential drug targets. To this end, we applied three different methods as follows:
First, we used the information supplied by STITCH (17), an aggregated database of interactions connecting >300 000 chemicals and 2.6 million proteins. We used the herbal ingredients’ general names and other alternative names to search STITCH and retrieved the related targets (protein); we then converted the corresponding target’s id into UniProt AC for unification purpose.
Second, the information from Herb Ingredients' Targets (HIT), which is extracted from published articles, was collected and integrated into our database.
Finally, as the information from HIT is mainly extracted from articles published in English, while the major TCM researches are in China, and the related research results are mainly published in Chinese, we collected these related articles published in Chinese and manually extracted the related information of ingredients and their targets from them. We used those herb names we collected and one of the following keywords ‘target’, ‘mechanism’, ‘pharmacology’ and ‘pharmacological’ to search Weipu database, which is like PubMed and is a system to host abstracts for the published articles in Chinese. Totally, we manually collected 680 herbal targets from >4500 articles. We also recorded the descriptions for the related experimental evidences and related URL or title for each article.
The six data fields in our database system are connected with their intrinsic relations (Figure 1): a prescription is composed of herbs, a herb contains various ingredients (compounds), an ingredient (or a drug) can interact with its targets (proteins) and a disease could be caused by the dysfunction of genes/proteins.
Figure 1.
Database structure. A–E: six data fields for prescription, herbs, ingredients, diseases, targets and drugs, respectively. 1–5: relationship used to connect each other. 1: prescription is composed of herbs. 2: herb contains ingredients. 3: ingredients can interact with targets. 4: drugs have identified targets. 5: targets may be the causes of disease.
DATABASE ACCESS AND NETWORK DISPLAY
Database query
As information and data from six different fields are connected, user can use any data filed to query the database and follow the link to retrieve related information. For example, user can choose herb as an entry point and use the herb’s name in English to conduct a query; the result page will display the queried herb’s information and show its connection to prescriptions and herbal ingredients, which will lead to the targets they interact with through those hyperlinks.
Network display
To virtually display the connection between herbs, herbal ingredients and their targets, we developed network-display tools, which provide more detailed network information. We also included protein–protein interaction in the network display to help user to check the potential combinational effect.
Herb–disease network
In TCM, herbs or prescriptions are formulated for particular ‘pattern’ (‘zheng’ in Chinese), which is a description of a specific functional state (18), whereas the drugs of modern medicine are designed for treating certain diseases. Therefore, it could be much useful to link the herbs or prescriptions to their treated diseases. As the disease-causing genes/proteins could be the targets of ingredients in a certain herb, we built such a herb–disease network based on this viewpoint (Figure 2).
Figure 2.
Herbal ingredient–target–disease network. Node: red triangle, herbal ingredient; blue circle, herbal ingredient’s target; yellow square, disease related to targets. Node size depends on its degree. Mouse over a node will display detailed information on the node.
Herbal ingredients—targets interaction network
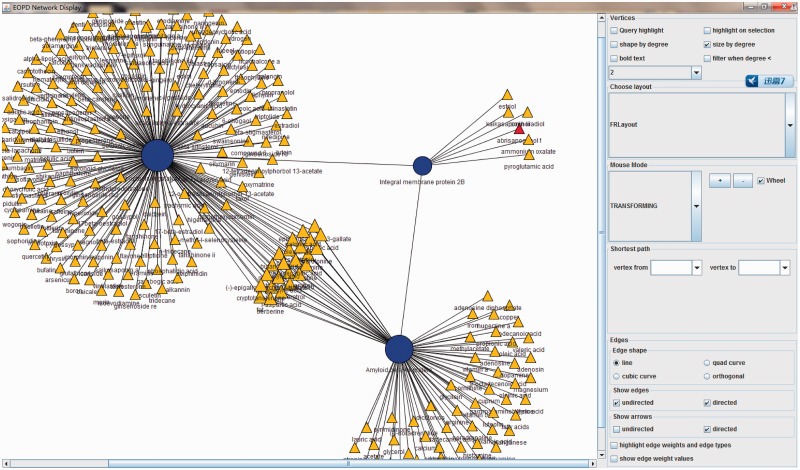
We generated this network to explore the interaction between ingredients and facilitate the study about combination therapy. To this end, we incorporated the protein–protein interactions from human protein reference database (19) into the network. In this network, proteins and ingredients are linked if those ingredients can target the proteins supported by either experiment evidence or computational methods. Therefore, based on the network, user can infer the potential synergistic/antagonistic effect between two ingredients if both ingredients can interact with the same protein or different proteins between which there exist interactions (Figure 3).
Figure 3.
Herbal ingredient–protein interaction network. Node: red triangle, the ingredient used for the query; yellow triangle, herbal ingredient(s); blue circle, herbal ingredient’s targets. Node size depends on its degree. Mouse over a node will display detailed information for the node.
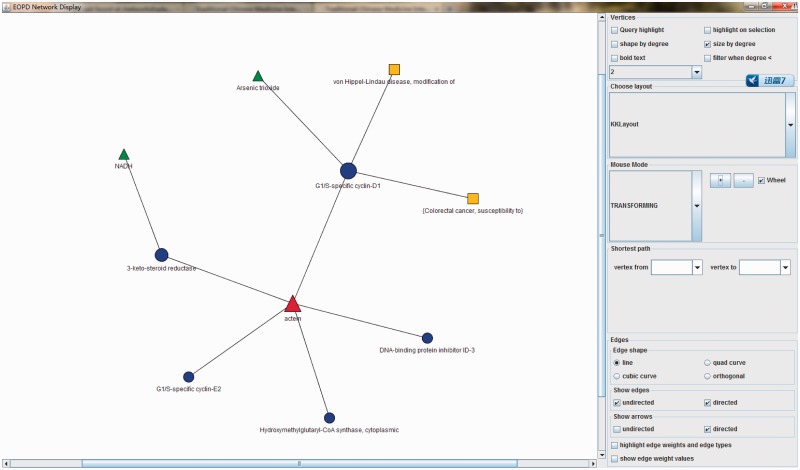
Herbal ingredient–target–disease–drug network
To explore the potential mechanisms underlying ingredients, we linked the ingredients to their potential targets, related diseases and approved drugs. Moreover, we built a tool to display the relationships in one network, which provides an intuitive view for user to infer the disease treatment mechanisms and identify potential ingredient’s targets through the connections between them. If a herbal ingredient can interact with a protein target, which is involved in a disease, it indicates a potential mechanism of the disease treatment for the ingredient. Additionally, if a herbal ingredient has the same protein target(s) as a drug, it implies a potential pharmacological effect for the ingredient (Figure 4).
Figure 4.
Herbal ingredient–target–disease–drug network. Node: red triangle, herbal ingredient; green triangle, drug; yellow square, disease; blue circle, herbal ingredient’s targets. Node size is related to its degree. Mouse over a node will display detailed information for the node.
To illustrate the usage of our database, we took Si-Wu-Tang (SWT) as an example. SWT is a famous TCM formula for treating menstrual discomfort and climacteric syndrome and is composed of four herbs, Radix Rehmanniae praeparata, Radix Angelicae Sinensis, Rhizoma Ligustici Chuanxiong and Radix Paeoniae Alba. Based on our database, we identified 30 ingredients from those herbs and their related targets; we also found that two drugs and 39 compounds from western medicine can link to SWT’s targets. For the two Food and Drug Administration-approved drugs, bevacizumab and ranibizumab, they are originally used to treat rectal cancer and diabetic retinopathy, respectively. Moreover, from our TCMID database, we also found that >20 TCM formulae have similar effects as SWT in treating menstrual discomfort and climacteric syndrome. All the formulae have common targets, caspase-3 and transcription factor AP-1, which are targeted by SWT as well. Therefore, it is suggested that those two targets would be potentially important therapeutical targets for the related diseases, and they would also be potential new targets for related drug discovery.
DISCUSSION
Our database system provides new insights of the mechanism study at molecular level for the TCM modernization. As TCM treats diseases in a more holistic way, it is necessary to adopt systematic methods to explore the potential mechanisms and therapeutic effect on TCM. Therefore, we tentatively bridge the modern western medicine and TCM through their common aspects—herbal ingredients/compounds with their targets, which makes the knowledge accumulated from >2000 years’ clinic practice and modern experience or computational methods meet together. This integrated information will not only benefit TCM’s moderations but also advance the development of network pharmacology.
It should be pointed out that many researches on herb ingredient targets are based on the activity measurements for particular enzymes or pathways, such as PDE-5 (ED), MRP1 (Leukemia) and casein kinase-2 (cancer). Therefore, those identified targets with biochemistry approaches may not be the real ones for those ingredients but are only affected by those herbs. However, the alterations of those gene expressions or the enzyme activities under herb treatments can provide implicitly information for exploring the real targets.
Many principles to describe the physiological conditions only belong to TCM, which seems mystery to western people, such as ‘Pattern’, ‘Zang-fu’ theory, ‘Qi’, ‘Xue’ and ‘Jinye’. These principles have great effect on the prescriptions or herbs selection or formulation, as herbs or prescriptions are formulated for certain ‘Patterns’ to make ‘Qi’, ‘Xue’ and ‘Jinye’ to return balance. Therefore, it is possible to link these mystery theories to proteins through herbs and helps to explain them at molecular level. For example, by collecting Qi-regulating herbs and connecting them to targets through herb–compound targets, we could figure out which pathway or module is related to ‘Qi’.
On the other hand, modern medicine researchers can also refer to our database for inspiration. It is well known that many herbs should be combined together to commit function, such as aconite and ginger, Chinese ephedra and cassia twig, which suggests that the ingredients of these pairs may have synergistic effect. Our databases provide information for such researches.
With the development of systems biology, an increasing number of ‘-omics’ methods, such as proteomics and metabonomics, are gradually adopted by TCM researches. Collecting such kind of information will definitely help promote the TCM’s systemic researches. Therefore, we intend to enclose microarray data, proteome data and other kind of data achieved from systemic methods into our database in the near future. Moreover, many folk prescriptions and traditional medicine are effective to treat certain rare or severe diseases (20), and collecting those formulae or medicines will be an urgent task, as many of them could be missed without prompted collection. Our database will be expanded to record this kind of information also.
ACKNOWLEDGEMENTS
The authors thank Mr. Chen Zhao for helping us to set up the web server and Mr. Peng Li for the network display design.
FUNDING
Funding for open access charge: National 973 Key Basic Research Program [2010CB945401, 2012CB910400]; National Natural Science Foundation of China [30870575, 31071162, 31000590, 81171272]; Science and Technology Commission of Shanghai Municipality [11DZ2260300]; “11th Five year plan” National Science and Technology Support Project [2007BAI20B06].
Conflict of interest statement. None declared.
REFERENCES
- 1.Qiu J. Traditional medicine—a culture in the balance. Nature. 2007;448:126–128. doi: 10.1038/448126a. [DOI] [PubMed] [Google Scholar]
- 2.Qiu J. China plans to modernize traditional medicine. Nature. 2007;446:590–591. doi: 10.1038/446590a. [DOI] [PubMed] [Google Scholar]
- 3.Normile D. Asian medicine. The new face of traditional Chinese medicine. Science. 2003;299:188–190. doi: 10.1126/science.299.5604.188. [DOI] [PubMed] [Google Scholar]
- 4.Wang MW, Hao X, Chen K. Biological screening of natural products and drug innovation in China. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 2007;362:1093–1105. doi: 10.1098/rstb.2007.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue TH, Roy R. Studying traditional Chinese medicine. Science. 2003;300:740–741. doi: 10.1126/science.300.5620.740. [DOI] [PubMed] [Google Scholar]
- 6.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 7.Kitano H. Towards a theory of biological robustness. Mol. Syst. Biol. 2007;3:137. doi: 10.1038/msb4100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitano H. Innovation—a robustness-based approach to systems-oriented drug design. Nat. Rev. Drug Discov. 2007;6:202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Zhou H, Liu YB, Wang JF, Li H, Ung CY, Han LY, Cao ZW, Chen YZ. Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br. J. Pharmacol. 2006;149:1092–1103. doi: 10.1038/sj.bjp.0706945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye H, Ye L, Kang H, Zhang DF, Tao L, Tang KL, Liu XP, Zhu RX, Liu Q, Chen YZ, et al. HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 2011;39:D1055–D1059. doi: 10.1093/nar/gkq1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CYC. TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. Plos One. 2011;6:e15939. doi: 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He M, Yan X, Zhou J, Xie G. Traditional Chinese medicine database and application on the Web. J. Chem. Inf. Comput. Sci. 2001;41:273–277. doi: 10.1021/ci0003101. [DOI] [PubMed] [Google Scholar]
- 13.Fang YC, Huang HC, Chen HH, Juan HF. TCMGeneDIT: a database for associated traditional Chinese medicine, gene and disease information using text mining. BMC Complement. Altern. Med. 2008;8:58. doi: 10.1186/1472-6882-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao XB, Hou TJ, Zhang W, Guo SL, Xu SJ. A 3D structure database of components from Chinese traditional medicinal herbs. J. Chem. Inf. Comput. Sci. 2002;42:481–489. doi: 10.1021/ci010113h. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Xie G, Yan X, editors. Encyclopedia of Traditional Chinese Medicines – Molecular Structures, Pharmacological Activities, Natural Sources and Applications. Vol. 6. New York: Springer; 2011. [Google Scholar]
- 16.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang WY. Therapeutic wisdom in traditional Chinese medicine: a perspective from modern science. Trends Pharmacol. Sci. 2005;26:558–563. doi: 10.1016/j.tips.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bang JY, Kim KS, Kim EY, Yoo HS, Lee YW, Cho CK, Choi Y, Jeong HJ, Kang IC. Anti-angiogenic effects of the water extract of HangAmDan (WEHAD), a Korean traditional medicine. Sci. China Life Sci. 2011;54:248–254. doi: 10.1007/s11427-011-4144-3. [DOI] [PubMed] [Google Scholar]