Abstract
Prediction of orthologs (homologous genes that diverged because of speciation) is an integral component of many comparative genomics methods. Although orthologs are more likely to have similar function versus paralogs (genes that diverged because of duplication), recent studies have shown that their degree of functional conservation is variable. Also, there are inherent problems with several large-scale ortholog prediction approaches. To address these issues, we previously developed Ortholuge, which uses phylogenetic distance ratios to provide more precise ortholog assessments for a set of predicted orthologs. However, the original version of Ortholuge required manual intervention and was not easily accessible; therefore, we now report the development of OrtholugeDB, available online at http://www.pathogenomics.sfu.ca/ortholugedb. OrtholugeDB provides ortholog predictions for completely sequenced bacterial and archaeal genomes from NCBI based on reciprocal best Basic Local Alignment Search Tool hits, supplemented with further evaluation by the more precise Ortholuge method. The OrtholugeDB web interface facilitates user-friendly and flexible ortholog analysis, from single genes to genomes, plus flexible data download options. We compare Ortholuge with similar methods, showing how it may more consistently identify orthologs with conserved features across a wide range of taxonomic distances. OrtholugeDB facilitates rapid, and more accurate, bacterial and archaeal comparative genomic analysis and large-scale ortholog predictions.
INTRODUCTION
The increase in genomic sequencing throughput has generated a rapid increase in the number of available bacterial and archaeal genome sequences (1,2). To make effective use of this growing resource, computational methods for comparative genomics analysis must keep pace, ideally without sacrificing accuracy or performance. Computationally predicted orthologs are integral to many comparative genomics analyses. Orthologs, related genes between species that have diverged as a result of speciation, are thought to more likely have similar functions than paralogs, which are homologous genes that have arisen through gene duplication (3). This ortholog functional conservation hypothesis or conjecture is the basis for many comparative genomics methods using computationally predicted orthologs to infer gene functions across species. Gene annotation transfer, phylogenetic profiling, metabolic network reconstruction and identification of gene regulatory elements by phylogenetic foot printing are examples of methods that use computationally predicted orthologs. Several platforms for comparative genomic analysis in microbial species have been developed, including the Comprehensive Microbial Resource (4), MicrobesOnline (5), Microbial Genome Database (6) and the Integrated Microbial Genomes database (7). Broadly, these resources specialize in data integration and functional inference based on comparative genomics. Some of these platforms could benefit from an ortholog prediction method that is both high throughput and highly precise. A number of high-throughput ortholog prediction methods have been developed. Although there are >30 ortholog databases, OMA (8), QuartetS (9,10), OrthoMCL (11,12), RoundUP (13), eggNOG (14) and HOGENOM (15,16) have associated web sites that provide ortholog predictions for significant numbers of microbial genomes (not necessarily specific to bacteria and archaea species). Ortholog prediction methods have been classified into two categories: tree-based methods, which use phylogenetic trees to resolve orthologs, and graph-based methods, which use pairwise sequence similarities computed across the entire genome (hybrid approaches have also been developed) (17). Shortcomings have been associated with both types (17). Tree-based methods require reliable automated tree building, accurate tree rooting and accurate species trees. Many of the methods do not scale well (17). Although graph-based methods typically scale better, as these methods often do not consider the broader phylogenetic context, they can generate false-positive ortholog predictions when the ortholog is missing (an ortholog can be missing because of incomplete genome annotations or gene deletions) (17,18). In the evaluations of ortholog prediction methods, the graph-based methods performed well, even in comparison with at least one sophisticated tree-based algorithm (19,20). The graph-based methods range significantly in their specificity and coverage. Some methods, such as the ortholog group-centric approaches: OrthoMCL, eggNOG and COG, have high coverage (producing many ortholog predictions), but lower specificity (higher false prediction rate) (20). Two methods have developed approaches to mitigate the misprediction due to missing ortholog genes. OMA and QuartetS are graph-based approaches that use additional outgroup genes to examine the broader phylogeny of a predicted ortholog pair to assess whether the predicted orthologs are more likely paralogs (8,9).
The reliability of the ortholog conjecture, that orthologs are more likely to have similar functions than paralogs, has recently been examined on a larger scale (21–26). These studies showed that for similar levels of divergence, orthologs tend to more often have functions that are more conserved than paralogs. The difference in function conservation, however, is not considerable and can vary between species and gene families (22,24–26). Although orthologs provide predictive power over paralogs when it comes to inferring genes functions across species, the implication from these studies is that ortholog prediction methods could be improved by targeting the set of orthologs that are functionally similar rather than all evolutionary orthologs.
Ortholuge is a high-throughput method that improves the specificity of ortholog prediction (18). It provides the benefits of graph-based methods, including scalability, but limits false positives generated by missing orthologs, as it considers the phylogenetic context of predicted orthologs. Ortholuge first predicts orthologs using the graph-based approach commonly called reciprocal best Basic Local Alignment Search Tool (BLAST) or RBB, but adds a second step where phylogenetic trees are built for each proposed orthologous gene/protein pair, rooted with a suitable outgroup. This phylogenetic analysis is completed for all predicted orthologs and, coupled with a statistical analysis (27), is used to flag orthologs that have diverged unusually versus what would be expected for the species. Many of the unusually diverging predicted orthologs are paralogs mispredicted as orthologs, or orthologs that have diverged more rapidly in one of the species (18). The remaining orthologs are more likely to have retained similar functions and may be better suited for many comparative genomic analyses. The phylogenetic approach used in Ortholuge does not require a separate species tree, making it especially suited for microbial genomes where species tree construction can be complicated by horizontal gene transfer and widely differing degrees of divergence between the species being compared. However, the Ortholuge method, initially made principally for our own internal use, has not been easily accessible. Ortholuge-based predictions for the human, mouse and cow genomes are available through InnateDB—a platform facilitating system-based analyses of the innate immune response (28)—and predictions for Pseudomonas species genomes are available through the Pseudomonas Genome Database (29). However, none of these predictions are queryable, and there is no flexibility in the display of ortholog prediction results. To make Ortholuge predictions widely available, we now report the development of OrtholugeDB, a web-accessible database of pre-computed Ortholuge-based predictions for completely sequenced bacterial and archaeal species (http://www.pathogenomics.sfu.ca/ortholugedb). However, OrtholugeDB is not limited to providing Ortholuge results. OrtholugeDB was also developed as a user-friendly and flexible interface for querying RBB-based orthologs predictions, in-paralog predictions (recently duplicated genes relative to the species being compared) and ortholog groups.
ORTHOLUGE
Ortholuge generates precise ortholog predictions between two species on a genome-wide scale using an additional outgroup genome for reference (18). It computes phylogenetic distance ratios for each pair of orthologs that reflect the relative rate of divergence for the predicted orthologs (Figure 1). Two ratios are needed to summarize the relative branch lengths for both ingroup genes. These phylogenetic ratios allow the user to distinguish between predicted orthologs with phylogenetic distance that is comparable with the species divergence [termed supporting-species-divergence (SSD) orthologs] and predicted orthologs with unusual divergence (non-SSD). SSD orthologs and orthologs undergoing unusual divergence have distinct ratio distributions. These ortholog types are observable when the ratios are plotted on a genome-wide scale (Figure 1D) (27). SSD and non-SSD ortholog assignments are determined by a statistical procedure that uses large-scale hypothesis testing approaches to infer the ratio distribution of the SSD orthologs and then assign a local false discovery rate (fdr) to each predicted ortholog pair based on this inferred distribution (27). The local fdr conveys the likelihood that a predicted ortholog pair is non-SSD given its ratio value (27). The implementation of Ortholuge, initially as a Perl pipeline, made it inaccessible to many researchers. We wanted to improve ease-of-access, and therefore, we constructed a database of pre-computed Ortholuge results. To scale the Ortholuge analysis, we automated the manual steps in the pipeline, including selecting outgroup species and computing ratio cutoffs for discriminating SSD orthologs from non-SSD orthologs.
Figure 1.
Overview of phylogenetic ratios used in Ortholuge. (A) Ratios computed by Ortholuge. The phylogenetic distances for the numerator (solid dark line) and the denominator (dashed dark line) are overlaid on top of the phylogenetic gene tree (light line) for two genes in the species of interest (Ingroup1 and Ingroup2) and a third reference species (Outgroup). Two ratios are needed to capture the proportional branch lengths of the Ingroup 1 and Ingroup 2 orthologs. (B) The RBB procedure can mistakenly identify a paralog as an ortholog when the true ortholog is missing (when the paralog forms an RBB with the remaining ortholog in the other species). Ortholuge can detect this case because the relative phylogenetic distance between the ingroup genes will increase causing ratio 1 value to become inflated [the numerator (solid dark line) and the denominator (dashed dark line) for ratio 1 are shown in the gene tree]. (C) An ortholog’s protein sequence can diverge more rapidly in one species versus another. This rapid sequence divergence can be associated with a change in function. Ortholuge detects cases where an ingroup gene’s relative phylogenetic divergence does not match the expected divergence for the species. (D) Histogram of ratio values. Plotting the distribution of ratio values reveals predicted orthologs whose relative phylogenetic divergence reflects species divergence (SSD) and predicted orthologs undergoing unusual divergence (non-SSD). The dashed lines represent ratio cutoffs. We also use an additional classification that we termed borderline-SSD to represent the ‘twilight’ region between SSD and non-SSD (region between dashed lines). (E) A ratio 1 × ratio 2 schematic showing how the combination of ratio 1 and ratio 2 assignments are used to assign a final Ortholuge classification for a predicted ortholog gene pair. Briefly, if the ratios provide different classifications, a lower value ratio’s classification is overridden by a higher value classification in the other ratio.
ORTHOLUGEDB
OrtholugeDB is a database of orthologs for bacteria and archaea. This database, which will be continually updated, provides Ortholuge-based ortholog predictions for completely sequenced bacterial and archaeal genomes from the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) database where a suitable outgroup is available. For those gene pairs where no suitable outgroup yet exists, RBB-based predictions are still presented. Ortholog predictions are available for protein-coding genes only [though predictions using DNA gene sequences are possible with the Ortholuge method (18)]. Data are stored in a MySQL database.
Content
OrtholugeDB is based on the completed genomes of bacterial and archaeal species from NCBI (incompletely sequenced genomes are not included in the database). The protein sequences for the bacterial and archaeal species were obtained using the MicrobeDB resource, a tool that provides a locally maintained database of sequenced microbial genomes from the NCBI Refseq database (1,2). In OrtholugeDB, the RBB procedure is used to generate the initial set of ortholog predictions (30). Multiple RBBs are possible, and we keep track of all of them, as these cases often represent recent gene duplications (in-paralogs). We evaluate the RBB-predicted orthologs using Ortholuge, and classify them as follows:
SSD: Predicted orthologs whose divergence (as reported by the Ortholuge phylogenetic ratios) is consistent with the divergence observed for the species. These predicted orthologs most likely represent valid orthologs, and have not undergone unusual divergence (such as accelerated evolution).
Borderline-SSD: Predicted orthologs with a phylogenetic ratio that is slightly higher than expected. When precision is critical to an application, these predicted orthologs can be excluded.
Divergent non-SSD: non-SSD genes have phylogenetic ratios that are significantly higher when compared with most other orthologs in the genomes [as per our statistical analysis, (27)], indicating that their divergence is not consistent with the species level of divergence. Based on our previous analyses/simulations (18), these are most likely incorrectly predicted orthologs, or orthologs that have undergone unusually rapid divergence because of a change in function.
Similar non-SSD: Similar non-SSDs have diverged unusually, as the length of one of the branches in the gene tree is proportionally longer than expected; however, the total phylogenetic distance separating the predicted orthologs is relatively small. Many Similar non-SSD genes will often be valid orthologs. However, the high phylogenetic ratio may suggest the genes are evolving at different rates.
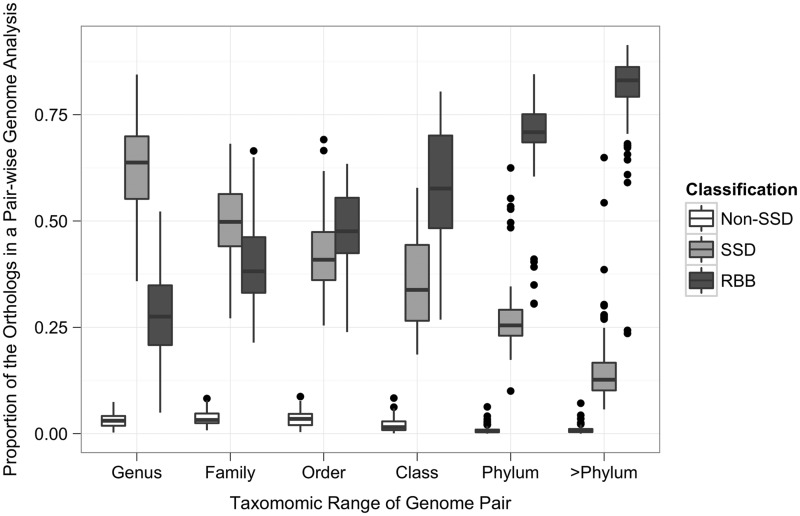
Boundaries between the Ortholuge classifications are based on the local fdrs described previously (27). Ortholuge requires an outgroup ortholog as a reference for computing the ratios (the outgroup gene is used to root the predicted ortholog’s phylogenetic tree). The ideal outgroup species diverged before the divergence of the comparison species, but also has a large number of common orthologs with the comparison species. The availability of suitable outgroups can impact the Ortholuge’s coverage. Species for which there are many closely related species’ genomes available tend to have better coverage. Evolutionary distance can also impact the number of possible Ortholuge assessments. As the species’ distance increases, the number of shared orthologs between the three species, the two comparison species and a more distantly related outgroup species, decreases. This is reflected in the proportion of RBB predictions in a genome-level analysis that are evaluated by Ortholuge among genomes that are evolutionarily more related compared with genomes that are evolutionarily distant. For species from the same genus, on average, 71.5% of RBB-based ortholog predictions are evaluated, but for species from different phyla, the average is 19.4%. Figure 2 shows the average proportion of RBB-predicted orthologs from a pairwise genome analysis that are evaluated by Ortholuge for species of various evolutionary distances. To select the reference outgroup species, we computed the optimum phylogenetic distance for an outgroup that best separates the distributions of SSD and non-SSD ratios for a given pair of ingroup species. The optimum distances formed the basis of a formula that we use to automatically select outgroups. Distances are computed using CVtree, a composition-based distance metric that reflects the evolutionary relatedness between species’ proteomes (31). In-paralogs are genes that have duplicated subsequent to species divergence. If the genes duplicated before the speciation (and creation of orthologs), the genes are referred to as out-paralogs. We identify in-paralogs using a procedure based on the InParanoid method (32). Briefly, after computing all orthologs, a gene is declared an in-paralog if it is closer to an ortholog in terms of BLAST score than the score between the orthologs.
Figure 2.
The proportion of the RBB-predicted orthologs from a pairwise genome analysis evaluated by Ortholuge for species with different evolutionary ranges. For each taxonomic level, 110 pairwise genomes were randomly selected, and the proportion of SSD, non-SSD and unevaluated orthologs (RBB) of the total number of orthologs predicted between that genome pair were calculated (species were selected based on the lowest common taxonomic grouping the species shared).
In addition to the pairwise orthologs, OrtholugeDB also contains pre-computed ortholog groups. These groups are transitive; all genes connected by an orthologous or in-paralogous relationship in the genomes under consideration are included in the group. We do post-processing of the groups to remove invalid ortholog connections. Incorrect ortholog connections can result in the fusion of separate ortholog groups. Most ortholog groups are densely connected. Incorrect ortholog predictions can appear as a limited number of edges bridging two densely connected subgroups (representing distinct ortholog groups). The post-processing step ensures that the groups maintain a certain overall level of connectivity by splitting groups along ortholog edges that have a normalized minimum cut value below the pre-defined threshold of 0.1 (a minimum cut is the number of edges needed to be removed to create two disjoint sub-graphs. A normalized minimum cut is the minimum cut value divided by the number of edges in the two resulting sub-graphs) (33). The tool Graclus is used to compute the normalized minimum cut for the groups (34).
Groups were constructed for multiple hierarchical levels representing sets of species with increasing phylogenetic distance. A hierarchical approach has a number of benefits. Ortholog prediction is more accurate between closely related species (35). Ortholog status is also relative to the species under consideration (i.e. an out-paralog can become an in-paralog when the depth of the last common ancestor of the species under investigation increases) (8,35). By providing ortholog groups for a number of levels, users can select their desired taxonomic range. The hierarchical levels in Ortholuge are based on CVtree distances (higher levels have a greater allowable CVtree distance) (31). Level distances were selected to match taxonomy classifications from the NCBI Taxonomy database (36,37). The benefit of CVtree distance-based levels is that the groups have a more consistent phylogenetic range than taxonomy classifications.
The method for computing OrtholugeDB ortholog groups is based on the hierarchical grouping approach developed for the OMA database (8) and the transitive grouping strategy used for computing Pseudomonas ortholog groups in the Pseudomonas Genome Database (38). There are, however, some differences between the approaches. The Pseudomonas Genome Database method is designed for computing ortholog groups at the genus level and uses conservation of gene order to resolve multiple RBB-predicted ortholog candidates (38). The use of gene order is not easily extended to species with broader phylogenetic distances; therefore, this step has been removed the OrtholugeDB procedure. In comparison to the OMA database method, differences include the use of normalized minimum cut in the OrtholugeDB approach versus the standard minimum cut value in the OMA approach to identify weak edges in the ortholog group connections. A standard minimum cut value tends to identify edges to small weakly connected subgroups (often the minimum cut in an ortholog group occurs along an ortholog edge linking a single gene). A normalized minimum cut value balances the minimum cut value with the connectivity in the two resulting partitions. Normalized minimum cuts can identify weak edges, which might not be the global minimum cut, but when removed produce densely connected subgroups (33). Another difference is the use of hierarchical groups based on CVtree distances in OrtholugeDB instead of the taxonomy classifications used in the OMA database (31,36,37). The CVtree distance thresholds in OrtholugeDB were selected to produce groups that are similar to the taxonomic groups. The immediate benefit of using a distance-based approach is that unclassified or incorrectly classified species will be properly grouped with species of similar distances. The other potential benefit of CVtree-based groupings is that they have a more consistent phylogenetic range than taxonomic classifications. One of the potential uses of hierarchical groups is to determine the timing of the emergence and disappearance of genes by tracking the genes in ortholog groups through multiple levels (8). Using groups based on a distance metric with equivalent phylogenetic ranges might provide the ability to more consistently compare the distribution of genes in diverse taxa.
Web interface
OrtholugeDB is designed to facilitate the rapid extraction and evaluation of bacterial and archaeal orthologs. Queries are intended to address a wide range of needs, from obtaining orthologs for single genes to orthologs for multiple genomes. OrtholugeDB includes the ability to run complex queries that filter genes based on the presence and absence of orthologs in other species (i.e. identifying genes unique to a species or set of species). The Ortholuge status of the predicted orthologs (SSD, borderline-SSD, non-SSD, etc.) is highlighted in the result pages. Results from any of the queries can be downloaded in tab-delimited, comma-separated values (CSV) and OrthoXML formats (39). The following types of queries are currently available in OrtholugeDB:
Orthologs between two genomes. Alternatively, you can return genes that do not have orthologs for one of the species (including in-paralogs). As an option, images showing the gene context for predicted orthologs can be generated (i.e. image displaying genes flanking the gene of interest).
Orthologs for a gene. The orthologs can be limited to a specified set of genomes or can be obtained for all species in the database. The gene context option is also available for this query.
Ortholog groups for a gene of interest. Groups for five hierarchical levels representing genus, family, order, class and phylum will be returned. To enhance viewing of the ortholog connections within the group, a graph view of the orthologs is provided. In the graph view, genes are viewed as nodes, and ortholog and in-paralog relationships are represented as edges between genes (Figure 3B). Ortholog edges are colored based on their Ortholuge status.
Compare the orthologs in a genome of interest across multiple other genomes. This query generates a high-level phyletic matrix view that quickly shows which genes in a genome of interest have orthologs in the specified comparison species (Figure 3A). Coding in the matrix highlights ortholog cardinality and Ortholuge status. Also provided as part this query is the ability to filter genes based on the presence or absence of orthologs in other species. This feature allows users to formulate complex queries, for example, obtaining genes that are common to one set of species (which may belong to divergent phyla but have a common phenotype), and not found in another set of species (with a different phenotype). A summary of the ortholog content is also provided for the query genome. The summary shows the proportion of protein-coding genes in the query genome that have no orthologs, one-to-one orthologous relationships or many-to-many orthologous relationships in each of the comparison species.
Figure 3.
Sample result views in OrtholugeDB. (A) The first few rows in the phyletic matrix view showing orthologs in Pseudomonas aeruginosa PAO1 and Burkholderia cenocepacia HI2424 and Burkholderia pseudomallei K96243. Viewing orthologs in 1–10 species is permitted. Numbering in cells indicates ortholog relationships without in-paralogs/co-orthologs (1:1), as well as genes in P. aeruginosa PAO1 that have no orthologs (0). Many-to-many, many-to-one, etc. are also possible values. Cells are colored based on their Ortholuge classification—blue: SSD orthologs, yellow: borderline-SSD, white: no Ortholuge classification available—are shown. (B) Graph view of the ortholog group associated with a putative copper resistance porin in Ralstonia pickettii 12J (GI: 187928805). The orthologs found at the genus hierarchical levels are shown (note: genome groupings in the hierarchical levels only approximate the taxonomic groups. Genomes were grouped using a consistent distance metric). Edge coloring between genes reflects the Ortholuge status and ortholog cardinality. Shown is an in-paralog relationship—represented by a pink edge—and an SSD or SSD ortholog—represented by a blue edge. Gray is the default edge color and represents an ortholog predicted by RBB.
Comparison of Ortholuge to other high-throughput ortholog prediction methods
Two other ortholog prediction methods use a reference genome to evaluate orthologs predicted by RBB: OMA (8) and QuartetS (9). OMA and QuartetS are high-throughput methods that produce pairwise ortholog predictions. In evaluations of the accuracy of the pairwise ortholog predictions, OMA was found to perform well in comparison with several other methods (20). QuartetS was shown to have a slightly lower false-positive rate than OMA (9). Similar to Ortholuge, QuartetS reconstructs a gene tree. However, QuartetS uses four genes, the predicted orthologs and two genes in a reference genome to build the gene tree, and instead of phylogenetic distances, QuartetS uses BLAST bit scores to represent branch lengths (9). Another significant difference is that QuartetS examines differences in the branch lengths, rather than ratios of the branch lengths (9). OMA also uses four genes to analyze predicted orthologs, but instead of reconstructing a gene tree, OMA uses heuristic rules to interpret the sequence information from the four genes (8). The approaches for selecting genes to use as references or outgroups are also different between OMA, QuartetS and Ortholuge. Ortholuge selects a single outgroup species and identifies potential orthologs in that species to use as reference genes (only a single species can be used because, to observe the divergence for all predicted orthologs relative to the species divergence, it must be on the same scale. If no ortholog exists, no evaluation of the ortholog is performed). OMA and QuartetS search all possible outgroup species for possible reference genes. This increases the number of RBB-predicted orthologs that are further analyzed in OMA and QuartetS.
A phylogenetic-based gold standard ortholog data set does not yet exist for microbial species. Typically, microbial ortholog data sets are evaluated by re-generating orthologs using computational methods that are deemed complementary to the original ortholog prediction method under evaluation (9,20), or by evaluating the functional similarity of the predicted orthologs using functional parameters such as manually annotated protein families and annotations (HAMAP, KEGG Orthology, GO) (9,20,40–43), or features of proteins and genes that are associated with function [protein domains and subcellular localization (SCL)] (20,40,44). Conservation of gene order in chromosomes is also used as a criterion to evaluate predicted orthologs, as conservation of gene neighborhood is often an indicator of conserved function among genes (20,40).
We compared OMA, QuartetS and Ortholuge methods by examining three criteria: similarity of protein domains, similarity of SCL, and whether the predicted orthologs displayed conserved gene order or synteny. Protein domains were obtained from InterPro database (45). SCLs were obtained from PSORTdb (46,47). An orthologous pair of genes was declared as being in a conserved gene region (displaying synteny) if at least one pair of adjacent genes were also orthologous. Although protein domains and SCL are highly conserved between orthologs, these features can also be conserved among closely related paralogs (40,48). As a result, using protein domain and SCL conservation as evaluation criteria will under-report the number of false positives. Similarly, gene synteny can occur between paralogs in cases where segmental duplications preserve the immediate gene neighborhood. To improve confidence in the inferred functional similarity, we looked at results produced from combining the three criteria. Results from individual criteria are available in Supplementary Figures S1–S3. For the combined analysis, we counted predicted orthologs having at least two of the three criteria: a domain conservation score >0.65 (44), the same localization and displaying synteny (based on the operational definition earlier) as a valid ortholog or true positive (note: because we are using indirect criteria and not a gold standard to assess performance, this is only an inferred true positive. Positive and negative numbers will likely be different in reality for the reasons stated earlier). Using this definition of positives and negatives, we compared precision, defined as TP/(TP + FP); recall, defined as TP/(TP + FN); accuracy, defined as (TP + TN)/(TP + TN + FP + FN) and Matthew’s coefficient constant (MCC), defined as:
for OMA, QuartetS and Ortholuge methods (Figure 4) (where TP = true positive, TN = true negative, FP = false positive and FN = false negative). For this evaluation, orthologs computed for 459 pairs of species were examined. The species pairs represented all taxonomic classifications and were equivalently distributed in all levels. Precision is the only metric available for OMA, as the OMA web site does not provide orthologs that were flagged as false by the method. MCC is an especially informative metric, as it is a balanced measure that reflects the correlation between true positives and negatives and the results from the prediction method (49). Methods that better segregate the positives and negatives will achieve higher MCC values. MCC values are higher for Ortholuge compared with QuartetS for a number of cutoffs. The difference in performance between Ortholuge and QuartetS (and OMA in terms of precision) is more pronounced for species with greater taxonomic separation. One possible explanation for this trend is that the features being examined, protein domains and SCL are less likely to be conserved among paralogs for distantly related species. Consequently, the number of false positives that are masked will be less for these species, suggesting that distantly related species might more accurately report the performance in this instance. Although clearly this is not an exhaustive evaluation of these methods (that would be beyond the scope of this article), Ortholuge, as implemented with the modifications described for more high-throughput analysis necessary for OrtholugeDB, is performing well. Overall, based on these criteria for evaluating orthologs functional similarity, Ortholuge appears to more consistently identify RBB-predicted orthologs with conserved features across a wide range of taxonomic distances.
Figure 4.
A comparison of the precision, recall, accuracy and MCC for the Ortholuge, QuartetS and OMA ortholog prediction methods using 459 pairwise bacterial and archaeal species analyses. In addition to computing performance values for all species combinations, genomes were organized according to the lowest taxonomic group that contained both species, and separate performance values were computed for genomes with different taxonomic ranges. Three cutoff values for the QuartetS method were examined. In this analysis, a ‘true positive’ is defined as having at least two of the following features as being conserved: SCL, protein domains or gene order. Negative results are not available for OMA; therefore, only precision was computed for the OMA method.
Comparison of the functionality in OrtholugeDB to OMA browser and QuartetS-DB
The ortholog prediction algorithms QuartetS and OMA have associated orthology databases: QuartetS-DB (10) and OMA Browser (8), respectively. They contain pairwise ortholog predictions and ortholog groups and are currently two of the largest orthology databases. OrtholugeDB was developed with a specific focus on ease-of-use and functionality. The queries and result views are designed for microbiologists to be able to (i) rapidly query NCBI genomes and identify shared or unique genes; (ii) access previously hard-to-obtain Ortholuge assessments when an outgroup is available; and (iii) visualize the orthologs for a gene or set of genomes. For querying shared and unique genes, the web interface in OrtholugeDB contains a flexible phyletic-based search that can identify common or unique genes in a genome of interest based on the presence or absence of orthologs in one or more comparison species. QuartetS-DB phyletic search is limited to identifying ortholog groups based only on the presence of orthologs in selected species, and OMA browser does not have phyletic-based search capabilities. OrtholugeDB also provides a separate rapid query for identifying the unique genes in a comparison of two genomes. OrtholugeDB includes a number of visualizations for ortholog data not offered in QuartetS-DB or OMA browser, such as the gene context view and the graph view of ortholog groups. The phyletic matrix view in OrtholugeDB efficiently displays the distribution of the genes in a query genome across multiple comparison genomes. Although QuartetS-DB has a phyletic-based tabular view of the ortholog groups, the phyletic matrix in OrtholugeDB is more informative, as it shows ortholog cardinality (i.e. one-to-one, many-to-many, no orthologs, etc.), plus Ortholuge classifications. The OrtholugeDB phyletic view also includes a summary page that shows the proportions of protein-coding genes in the query genome associated with each type of orthologous relationship. Gene duplications are important to consider when inferring which orthologs are likely to have similar functions. If a particular ortholog has duplicated in a genome, it is not straightforward to determine how the gene function is impacted (for example, whether one of the duplicated genes maintained the ancestral gene function or sub-functionalization between the duplicated genes occurred) (50). In-paralog predictions are integrated into the result views in OrtholugeDB to flag ortholog genes that have undergone duplication after species divergence. In QuartetS-DB, in-paralog groups must be obtained through a separate query.
CONCLUSIONS
Ortholuge, by identifying orthologs that diverged to the same relative degree as their species, produces a set of orthologs that are more likely to have retained similar function and are better suited for comparative genomic analyses. OrtholugeDB makes Ortholuge analysis readily available for bacterial and archaeal species and where Ortholuge assessments are not available provides RBB-based ortholog predictions. The OrtholugeDB web site is designed to facilitate the retrieval of orthologs for single genes to multiple genomes. It includes features that allow high-level visualization of orthologs and formulating complex queries to retrieve orthologs. OrtholugeDB facilitates bacterial and archaeal comparative genomic analysis by providing large-scale ortholog predictions, a flexible search interface and further more precise assessment of orthologs when suitable reference genomes are available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Genome Canada; Genome BC; Natural Sciences and Engineering Research Council of Canada (NSERC); Cystic Fibrosis Foundation. Funding for open access charge: NSERC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Raymond Lo and other members of the Brinkman laboratory for their review of this manuscript and OrtholugeDB suggestions. They also thank the NCBI and all those institutions that made their genome sequence data publicly accessible. M.D.W. was a Michael Smith Foundation for Health Research (MSFHR) trainee, and F.S.L.B. was a MSFHR Senior Scholar.
REFERENCES
- 1.Langille MGI, Laird MR, Hsiao WWL, Chiu TA, Eisen JA, Brinkman FSL. MicrobeDB: a locally maintainable database of microbial genomic sequences. Bioinformatics. 2012;28:1947–1948. doi: 10.1093/bioinformatics/bts273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 4.Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinsky K, White O, et al. The comprehensive microbial resource. Nucleic Acids Res. 2010;38:D340–D345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010;38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchiyama I, Higuchi T, Kawai M. MBGD update 2010: toward a comprehensive resource for exploring microbial genome diversity. Nucleic Acids Res. 2010;38:D361–D365. doi: 10.1093/nar/gkp948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–D294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu C, Zavaljevski N, Desai V, Reifman J. QuartetS: a fast and accurate algorithm for large-scale orthology detection. Nucleic Acids Res. 2011;39:e88. doi: 10.1093/nar/gkr308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C, Desai V, Cheng L, Reifman J. QuartetS-DB: a large-scale orthology database for prokaryotes and eukaryotes inferred by evolutionary evidence. BMC Bioinformatics. 2012;13:143. doi: 10.1186/1471-2105-13-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLuca TF, Cui J, Jung J-Y, St Gabriel KC, Wall DP. Roundup 2.0: enabling comparative genomics for over 1800 genomes. Bioinformatics. 2012;28:715–716. doi: 10.1093/bioinformatics/bts006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufayard J-F, Duret L, Penel S, Gouy M, Rechenmann F, Perrière G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- 16.Penel S, Arigon A-M, Dufayard J-F, Sertier A-S, Daubin V, Duret L, Gouy M, Perrière G. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009;10(Suppl. 6):S3. doi: 10.1186/1471-2105-10-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzniar A, van Ham RCHJ, Pongor S, Leunissen JAM. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Fulton DL, Li YY, Laird MR, Horsman BGS, Roche FM, Brinkman FS. Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics. 2006;7:270. doi: 10.1186/1471-2105-7-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulsen T, Huynen MA, de Vlieg J, Groenen PMA. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nehrt NL, Clark WT, Radivojac P, Hahn MW. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput. Biol. 2011;7:e1002073. doi: 10.1371/journal.pcbi.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forslund K, Pekkari I, Sonnhammer ELL. Domain architecture conservation in orthologs. BMC Bioinformatics. 2011;12:326. doi: 10.1186/1471-2105-12-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas PD, Wood V, Mungall CJ, Lewis SE, Blake JA. On the use of gene ontology annotations to assess functional similarity among orthologs and paralogs: a short report. PLoS Comput. Biol. 2012;8:e1002386. doi: 10.1371/journal.pcbi.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altenhoff AM, Studer RA, Robinson-Rechavi M, Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 2012;8:e1002514. doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dessimoz C, Gabaldón T, Roos DS, Sonnhammer ELL, Herrero J. Toward community standards in the quest for orthologs. Bioinformatics. 2012;28:900–904. doi: 10.1093/bioinformatics/bts050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson ME, Chen F, Saven JG, Roos DS, Babbitt PC, Sali A. Evolutionary constraints on structural similarity in orthologs and paralogs. Protein Sci. 2009;18:1306–1315. doi: 10.1002/pro.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min JE, Whiteside MD, Brinkman FSL, McNeney B, Graham J. A statistical approach to high-throughput screening of predicted orthologs. Comput. Stat. Data Anal. 2011;55:935–943. [Google Scholar]
- 28.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan THW, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winsor GL, Lam DKW, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock REW, Brinkman FSL. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2010;39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, Hao B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009;37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostlund G, Schmitt T, Forslund K, Köstler T, Messina DN, Roopra S, Frings O, Sonnhammer ELL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi J, Malik J. Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000;22:888–905. [Google Scholar]
- 34.Dhillon IS, Guan Y, Kulis B. Weighted graph cuts without eigenvectors a multilevel approach. IEEE Trans. Pattern Anal. Mach. Intell. 2007;29:1944–1957. doi: 10.1109/TPAMI.2007.1115. [DOI] [PubMed] [Google Scholar]
- 35.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock REW, Brinkman FS. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009;37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt T, Messina DN, Schreiber F, Sonnhammer ELL. Letter to the editor: SeqXML and OrthoXML: standards for sequence and orthology information. Brief. Bioinform. 2011;12:485–488. doi: 10.1093/bib/bbr025. [DOI] [PubMed] [Google Scholar]
- 40.Hulsen T, Huynen MA, de Vlieg J, Groenen PMA. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rey S, Acab M, Gardy JL, Laird MR, deFays K, Lambert C, Brinkman FSL. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 2005;33:D164–D168. doi: 10.1093/nar/gki027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu NY, Laird MR, Spencer C, Brinkman FSL. PSORTdb—an expanded, auto-updated, user-friendly protein subcellular localization database for Bacteria and Archaea. Nucleic Acids Res. 2011;39:D241–D244. doi: 10.1093/nar/gkq1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nair R, Rost B. Sequence conserved for subcellular localization. Protein Sci. 2002;11:2836–2847. doi: 10.1110/ps.0207402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vihinen M. How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics. 2012;13(Suppl. 4):S2. doi: 10.1186/1471-2164-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]