Abstract
Here, we present WormQTL (http://www.wormqtl.org), an easily accessible database enabling search, comparative analysis and meta-analysis of all data on variation in Caenorhabditis spp. Over the past decade, Caenorhabditis elegans has become instrumental for molecular quantitative genetics and the systems biology of natural variation. These efforts have resulted in a valuable amount of phenotypic, high-throughput molecular and genotypic data across different developmental worm stages and environments in hundreds of C. elegans strains. WormQTL provides a workbench of analysis tools for genotype–phenotype linkage and association mapping based on but not limited to R/qtl (http://www.rqtl.org). All data can be uploaded and downloaded using simple delimited text or Excel formats and are accessible via a public web user interface for biologists and R statistic and web service interfaces for bioinformaticians, based on open source MOLGENIS and xQTL workbench software. WormQTL welcomes data submissions from other worm researchers.
INTRODUCTION
Over the past 30 years, the metazoan Caenorhabditis elegans has become a premier animal model for determining the genetic basis of quantitative traits (1,2). The extensive knowledge of molecular, cellular and neural bases of complex phenotypes makes C. elegans an ideal system for the next endeavour: determining the role of natural genetic variation on system variation. These efforts have resulted in an accumulation of a valuable amount of phenotypic, high-throughput molecular and genotypic data across different developmental worm stages and environments in hundreds of strains (3–19). In addition, a similar wealth has been produced on hundreds of different C. elegans wild isolates and other species (20). For example, C. briggsae is an emerging model organism that allows evolutionary comparisons with C. elegans and quantitative genetic exploration of its own unique biological attributes (21).
This rapid increase in valuable data calls for an easily accessible database allowing for comparative analysis and meta-analysis within and across Caenorhabditis species (22). To facilitate this, we designed a public database repository for the worm community, WormQTL (http://www.wormqtl.org). Driven by the PANACEA project of the systems biology program of the EU, its design was tuned to the needs of C. elegans researchers via an intensive series of interactive design and user evaluation sessions on a mission to integrate all available data within the project.
As a result, data that were scattered across different platforms and databases can now be stored, downloaded, analysed and visualized in an easily and comprehensive way in WormQTL. On top, the database provides a set of user interfaced analysis tools to search the database and explore genotype–phenotype mapping based on R/qtl (23,24). New data can be uploaded and downloaded using the extensible plain text format for genotype and phenotypes, XGAP (25). There is no limit to the type of data (from gene expression to protein, metabolite or cellular data) that can be accommodated because of its extensible design. All data and tools can be accessed via a public web user interface and programming interfaces to R and REST web services, which were built using the MOLGENIS biosoftware toolkit (26). Moreover, users can upload and share more R scripts as ‘plugin’ for the colleagues in the community to use directly and run those on a computer cluster using software modules from xQTL workbench (27); this requires login to prevent abuse. All software can be downloaded for free to be used, for example as local mirror of the database, and/or to host new studies.
All the software was built as open source, reusing and building on existing open source components as much as possible. WormQTL is freely accessible without registration and is hosted on a large computational cluster enabling high-throughput analyses at http://www.wormqtl.org. Below we detail the results, methods used to implement the system and future plans.
RESULTS
WormQTL is an online database platform for expression quantitative trait loci (eQTL) exploration to service the worm community and already provides many publicly available data sets (5,9–15,19). New data sets can be uploaded using the XGAP plain file data format. Suitable help pages are provided. Currently, 38 public data sets have been loaded, of which the bulk is xQTL data on 500 strains (introgression lines, recombinant inbred lines (RILs), recombinant inbred advanced intercross lines and natural isolates), 55,000 transcripts, 1594 samples and 1579 markers (Table 1). With this combination of classical phenotypes, molecular profiles and genetics data sets, WormQTL contains all the ‘genetical genomics’ experiments published to our current knowledge (except for some tiling data). Using WormQTL, researchers can explore many xQTLs across the various studies in different conditions and ages and compare classical QTLs with xQTLs. The main interfaces are ‘Find QTLs’, ‘Genome browser’ and‘Browse data’.
Table 1.
Overview of data sets currently loaded
| Phenotypes | Sample size | Parental strains | Reference | Pubmed link | Growing temperature | Stage |
|---|---|---|---|---|---|---|
| Gene expression | 2 × 40 RILs | CB4856; N2 | Li et al. (10) | 17196041 | 16 and 24°C | (72 h at 16 and 40 h at 24) L4 |
| Gene expression | 60 RILs | CB4856; N2 | Li et al. (11) | 20610403 | 24°C | (40 h) L4 |
| Gene expression | 36 × 3 RILs | CB4856; N2 | Viñuela et al. (14) | 20488933 | 24°C | (40, 96 and 214 h) L4, adult, old |
| Gene expression | 208 RIAILs | CB4856; N2 | Rockman et al. (5) | 20947766 | 20°C | YA |
| Feeding curves RNAi exposure | 56 RILs × 12 RNAi | CB4856; N2 | Elvin et al. (15) | 22004469 | 20°C | Multi-generational |
| Life-history traits | 80 RILs | CB4856; N2 | Gutteling et al. (13) | 16955112 | 12 and 24°C | Egg, L4, YA |
| Lifespan and pharyngeal-pumping | 90 NILs | CB4856; N2 | Doroszuk et al. (9) | 19542186 | 20°C | All; synchronized |
| Lifespan, Recovery and reproduction after heat-shock | 58 RILs | CB4856; N2 | Rodriguez et al. (19) | 22613270 | 20 and 35°C heat shock | L4 and adult |
| Gene expression | 6 × 2 parental strains | CB4856 and N2 | Viñuela et al. (18) | 22670229 | 24°C | (40, 96 and 214 h) L4, adult, old |
RILs, recombinant inbred lines; NILs, near isogenic lines; RIAILs, recombinant inbred advanced intercross lines.
Find QTLs
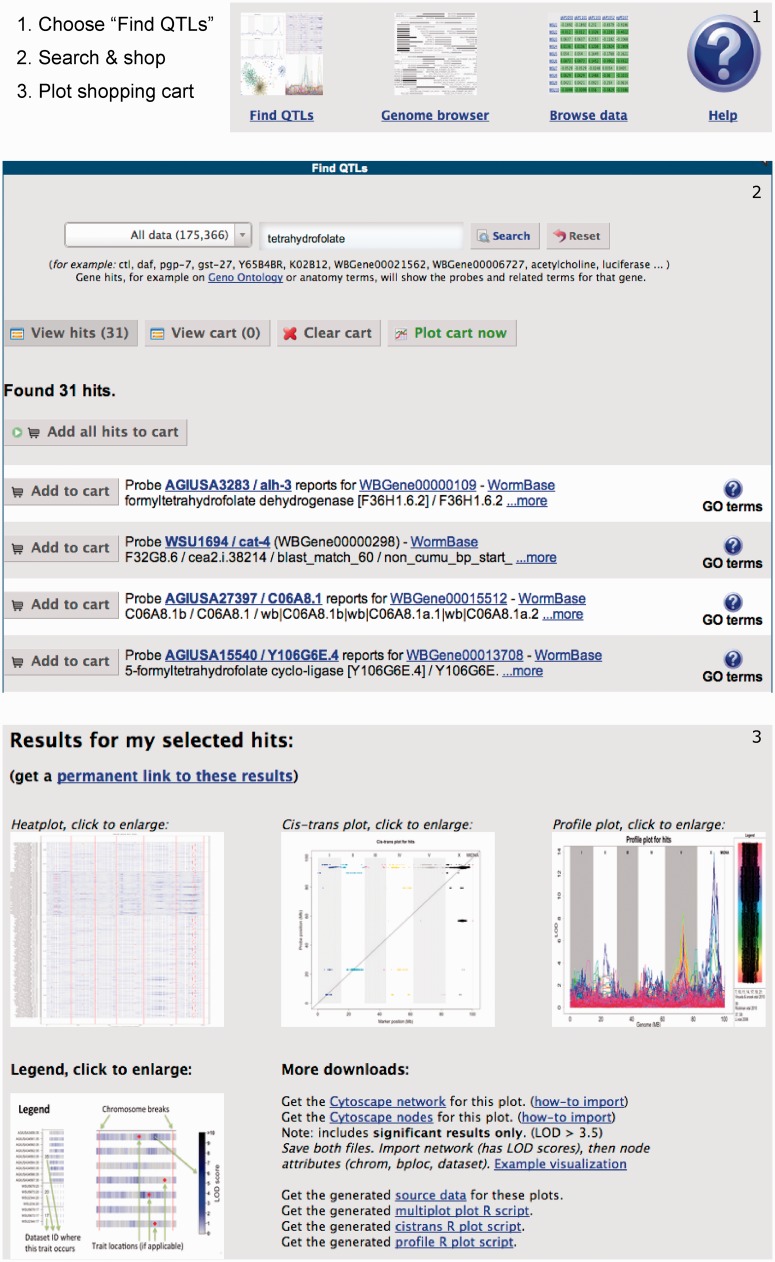
QTL is genomic regions associated with phenotypic variation and can be used to study the genetic architecture of traits and to detect potential phenotypic regulators. Recently, the number of QTLs and especially eQTL studies in C. elegans has increased greatly. These eQTL studies consist of large data sets that, before WormQTL, were very difficult to access and perform a combined meta-analysis. Therefore, we provide easy access to most of the eQTL studies published, by search, browse and plot functions (Figure 1).
Figure 1.
Cross experiment search. (1) Users can search for genes, markers or traits of interest using a google-like search box, optionally filtering for particular types of information. The results include links to WormBase and PubMed where possible. (2) From the resulting list, users can select items in a shopping cart, optionally repeating the search to add more items. (3) Finally, users can plot the contents of the shopping cart on top of all collected QTL data sets showing interesting areas in a heat plot, significant traits in a cis/trans plot and the individual signals in a profile plot. Alternatively, users can browse the QTL profiles using a genome browser, view/download the data set by simply browsing trough all available information or use the scriptable interface to program against. A complete tutorial is available in the help page.
We support relatively simple questions like ‘does my gene have an xQTL?’ to more advanced ones like ‘how do these genes fit into an xQTL network?’. All the matching genes, markers and traits found in the data sets are returned including links to WormBase and literature. Furthermore, WormQTL is the first portal for any species that allows comparison of eQTLs over multiple experiments and environments, giving insight in the plastic nature of genetic regulation.
Genome browser
To find the genes that have a QTL on your favourite position, click ‘Genome browser’. Here, you can select from all the different releases of the University of California, Santa Cruz genome releases. You can add tracks from the designated experiments of interest. Then navigate to your favourite location (tip: use open in new window) and collect significant probe identifiers from that region.
Browse data
Complete data sets and accompanying gene, sample and trait identifier lists can be browsed via the ‘browse data’ user interface. External identifiers anywhere in the data are automatically recognized and enhanced as linkouts to background information, such as links to Wormbase, NCBI, KEGG or Ensembl. All the annotation lists and data matrices can be browsed and searched in a tabular form and can be downloaded as plain text or Excel files. Readers can also download data sets or submit new data sets using the XGAP data format following examples described in the WormQTL help section. Also all data can be accessed programmatically from with R (as whole matrix or per row) or using REST web services, including filtering of the annotations (genes, probes, markers and phenotypes) and services to ‘slice’ individual lines out of the complete data sets to speed up download and (parallel) analyses. Alternatively, readers can request a login to upload data and new analysis scripts directly.
DISCUSSION
Implementation
All the software was implemented using the open source ‘Molecular Genetics Information Systems’ MOLGENIS toolkit (26), and in particular one previously existing MOLGENIS application, the extensible xQTL workbench (27) and the R/qtl QTL mapping and visualization package for the R language (23,24). The MOLGENIS toolkit is a Java-based software to generate tailored research infrastructure on demand (22). From a single ‘blueprint’ describing all biological data structures and user interfaces of the whole system, MOLGENIS autogenerates a full application including user interface, database infrastructure and application programming interfaces (APIs) in R, REST and SOAP.
At the push of a button, MOLGENIS ‘generators’ automatically translates these models into a database, standard user interfaces for data queries and updates, upload/download tools for tab-delimited data and scriptable interfaces for programmers to users from within R and via web services. This greatly speeded up the initial software development and also enables rapid extension when, for example, new data types arrive. On top of this foundation, we build the WormQTL specific user interactions such as the ‘Find QTLs’ and the ‘Genome browser’ using MOLGENIS ‘plug-in’ mechanism and the visualizations and plots using the R interface. xQTL workbench is a scalable web platform for the mapping of QTLs at multiple levels: for example, gene expression (xQTL), protein abundance (pQTL), metabolite abundance (mQTL) and phenotype (phQTL) data. The xQTL workbench provided a set of previously developed user interfaces to run R/qtl mapping methods directly from within the WormQTL user interface, the ability to add new analysis procedures in R, data management and data format conversions, all greatly speeding up the generation of new xQTL profiles.
All the data sets were downloaded from their original sources and then formatted using the XGAP data format. XGAP is a simple text file format that uses a directory of tab-delimited files or one Excel file with multiple sheets to load lists of annotations and data matrices. The annotations list all the background information needed to run and interpret the analysis including, for example, genome position information, such as markers, genes, probes and strains. The data matrices describe all the raw, intermediate and result data, such as gene expression, genotypes and QTL P-values, with the row names and column names cross linking to the annotations. For example, gene expression is a matrix of ‘gene’ X ‘sample’. Subsequently these data sets were loaded using the MOLGENIS/xQTL data import wizards, which check the files for correctness and give informative feedback if the data are not yet in a format that WormQTL can understand (25). All the annotations are stored in tables in the database; the large data matrices are stored in a optimized binary format to speed up analyses and queries. This format is documented in the WormQTL manual to ease the submission of new data sets from the community. Finally, all the QTL profiles were recalculated according to the specification of the original, or slightly modified when needed, such as to include a previously missing wrongly labelled sample correction. In this process, we greatly benefitted from the integration with xQTL workbench, which enabled us to re-run all these analyses on the computer cluster and add new R analysis procedures when needed, simply from the user interface.
All software is available as open source on http://github.com/molgenis for others to reuse locally, and related technical documentation is available at http://www.xqtl.org and http://www.rqtl.org and http://www.molgenis.org.
Future plans
The current version of WormQTL (June 2012) is a comprehensive, versatile and flexible package. Follow-up plans of more extended versions with new tools and data depend on the demand by the users of WormQTL. We envisage that in the future, three types of new tools will be developed: (i) visualization tools, (ii) QTL mapping tools and (iii) candidate gene selection tools. Improved visualization tools might include plotting a phenotype against the marker at a certain position; so the two groups become visible at a QTL position. Also plots can be made showing transgression and heritability per microarray probe or gene or histograms of the phenotypic values (and include the parental values if available). Advanced QTL mapping tools might include multi-environment/age mapping or genotype-by-environment analyses, developed in collaboration with the R/qtl team to enable automatic links to this software. The candidate gene selection tools would benefit from the most recent stable release of Wormbase (28), the most widely used platform for worm biology. But also other sources of information like MODENCODE (29) or Wormnet (30) are likely to be connected with WormQTL. A candidate gene selection tool might be implemented in a next version of WormQTL as it is less easy to implement and often needs information beyond WormQTL. One can think of (i) which SNPs/genes/polymorphic genes/transcription factor binding sites and so forth are underlying a eQTL; (ii) which gene, underlying my xQTL, is linked to most of the genes having an xQTL; (iii) which genes are polymorphic and (iv) which other genotypes show a difference in expression and do they share polymorphisms with the parental strains of the RIL population that the xQTL was mapped in. Moreover, WormQTL can be easily expanded to other Caenorhabditis species (21).
We believe that WormQTL, which will be continuously curated by the members of this international consortium, is a very attractive database for the growing community of quantitative genetics in worms researchers. We are committed to maintain data and software for the years to come and invite the community to add and share new data and ideas.
FUNDING
The Centre for BioSystems Genomics (CBSG) and the Netherlands Consortium of Systems Biology (NCSB), both of which are part of the Netherlands Genomics Initiative of the Netherlands Organisation for Scientific Research (NWO) (to D.A.); European Community's Health Seventh Framework Programme (FP7/2007-2013) under grant agreement PANACEA [222936 to L.B.S., M.E., T.S., J.E.K., R.C.J.]; ERASysbio-plus ZonMW project GRAPPLE - Iterative modelling of gene regulatory interactions underlying stress, disease and ageing in C. elegans [90201066 to L.B.S.]. Funding for open access charge: EU 7th Framework Programme under the Research Project PANACEA (no: 222936).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Wormbase for being an easy accessible and versatile data source. We also thank Mark Sterken and Rita Volkers for helpful suggestions, Konrad Zych for graphics design and testing and many members of the C. elegans community for their comments and ideas.
REFERENCES
- 1.Gaertner B, Phillips PC. Caenorhabditis elegans as a platform for molecular quantitative genetics and the systems biology of natural variation. Genet. Res. 2010;92:331–348. doi: 10.1017/S0016672310000601. [DOI] [PubMed] [Google Scholar]
- 2.Kammenga JE, Phillips PC, de Bono M, Doroszuk A. Beyond induced mutants: using worms to study natural variation in genetic pathways. Trends Genet. 2008;24:178–185. doi: 10.1016/j.tig.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, And Kruglyak L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kammenga JE, Doroszuk A, Riksen JAG, Hazendonk E, Spiridon L, Petrescu A-J, Tijsterman M, Plasterk RHA, Bakker J. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 2007;3:0358–0366. doi: 10.1371/journal.pgen.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330:372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doroszuk A, Snoek LB, Fradin E, Riksen JAG, Kammenga JE. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 2009;37:e110. doi: 10.1093/nar/gkp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JAG, Hazendonk E, Prins P, Plasterk RH, Jansen RC, et al. Mapping determinant of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:2155–2161. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Breitling R, Snoek LB, van der Velde KJ, Swertz MA, Riksen J, Jansen RC, Kammenga JE. Global genetic robustness of the alternative splicing machinery in Caenorhabditis elegans. Genetics. 2010;186:405–410. doi: 10.1534/genetics.110.119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutteling EW, Doroszuk A, Riksen JAG, Prokop Z, Reszka J, Kammenga JE. Environmental influence on the genetic correlations between life-history traits in Caenorhabditis elegans. Heredity. 2007;98:206–213. doi: 10.1038/sj.hdy.6800929. [DOI] [PubMed] [Google Scholar]
- 13.Gutteling EW, Riksen JAG, Bakker J, Kammenga JE. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity. 2007;98:28–37. doi: 10.1038/sj.hdy.6800894. [DOI] [PubMed] [Google Scholar]
- 14.Viñuela A, Snoek LB, Riksen JAG, Kammenga JE. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 2010;20:929–937. doi: 10.1101/gr.102160.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elvin M, Snoek LB, Frenjo M, Klemstein U, Kammenga JE, Poulin GB. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genomics. 2011;12:510. doi: 10.1186/1471-2164-12-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler CH. Cryptic intraspecific variation in sex determination in Caenorhabditis elegans revealed by mutations. Heredity. 2010;105:473–482. doi: 10.1038/hdy.2010.62. [DOI] [PubMed] [Google Scholar]
- 17.Harvey SC, Shorto A, Viney ME. Quantitative genetic analysis of life-history traits of Caenorhabditis elegans in stressful environments. BMC Evol. Biol. 2008;8:15. doi: 10.1186/1471-2148-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viñuela A, Snoek LB, Riksen JAG, Kammenga JE. Aging uncouples heritability and expression-QTL in Caenorhabditis elegans. G3. 2012;2:597–605. doi: 10.1534/g3.112.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez M, Snoek LB, Riksen JAG, Bevers RP, Kammenga JE. Genetic variation for stress-response hormesis in C. elegans lifespan. Exp. Geront. 2012;47:581–587. doi: 10.1016/j.exger.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Felix MA, Kruglyak L. Recent chromosome-scale selective sweeps reshaped genomic diversity in C. elegans. Nat. Genet. 2012;29:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross JA, Koboldt DC, Staisch JE, Chamberlin HM, Gupta BP, Miller RD, Baird SE, Haag ES. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 2011;7:e1002174. doi: 10.1371/journal.pgen.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swertz MA, Jansen RC. Beyond standardization: dynamic software infrastructures for systems biology. Nat. Rev. Genet. 2007;8:235–243. doi: 10.1038/nrg2048. [DOI] [PubMed] [Google Scholar]
- 23.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 24.Arends D, Prins P, Jansen RC, Broman KW. R/qtl: high throughput multiple QTL Mapping. Bioinformatics. 2010;26:2990–2992. doi: 10.1093/bioinformatics/btq565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swertz MA, van der Velde KJ, Tesson BM, Scheltema RA, Arends D, Vera G, Dijkstra M, Schofield P, Schughart K, Hancock JM, et al. XGAP: a uniform and extensible data model and software platform for genotype and phenotype experiments. Genome Biol. 2010;11:R27. doi: 10.1186/gb-2010-11-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swertz MA, Dijkstra M, Adamusiak T, van der Velde KJ, Kanterakis A, Roos ET, Lops J, Thorisson GA, Byelas G, Muilu J, et al. The MOLGENIS toolkit: rapid prototyping of biosoftware at the push of a button. BMC Bioinformatics. 2010;11(Suppl. 12):S12. doi: 10.1186/1471-2105-11-S12-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arends D, van der Velde KJ. xQTL workbench: a scalable web environment for multilevel QTL analysis. Bioinformatics. 2012;28:1042–1044. doi: 10.1093/bioinformatics/bts049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yook K, Harris TW, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, de la Cruz N, Duong A, Fang R, et al. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 2012;40:D735–D741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat. Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]