Abstract
The subcellular location database for Arabidopsis proteins (SUBA3, http://suba.plantenergy.uwa.edu.au) combines manual literature curation of large-scale subcellular proteomics, fluorescent protein visualization and protein–protein interaction (PPI) datasets with subcellular targeting calls from 22 prediction programs. More than 14 500 new experimental locations have been added since its first release in 2007. Overall, nearly 650 000 new calls of subcellular location for 35 388 non-redundant Arabidopsis proteins are included (almost six times the information in the previous SUBA version). A re-designed interface makes the SUBA3 site more intuitive and easier to use than earlier versions and provides powerful options to search for PPIs within the context of cell compartmentation. SUBA3 also includes detailed localization information for reference organelle datasets and incorporates green fluorescent protein (GFP) images for many proteins. To determine as objectively as possible where a particular protein is located, we have developed SUBAcon, a Bayesian approach that incorporates experimental localization and targeting prediction data to best estimate a protein’s location in the cell. The probabilities of subcellular location for each protein are provided and displayed as a pictographic heat map of a plant cell in SUBA3.
INTRODUCTION
The sequencing of the genome of the model plant Arabidopsis thaliana (1) and the subsequent development of extensive tools and datasets for its genetic dissection (2,3) has provided scientists with foundational information on the structure of model plant genomes and their coding capacities. However, the function of most Arabidopsis proteins still remains to be resolved. A key step towards understanding the metabolic or biochemical role of any protein is to define its subcellular location. Proteins found in distinct subcellular compartments are part of interconnected metabolic and regulatory pathways, can share similar characteristics and collectively define the function of the particular compartment. Aggregating the evidence for where all the proteins of Arabidopsis are located in cells is thus an important foundation for interpreting the role of each of its genes (4).
Both in silico prediction methods and experimental approaches are widely used by researchers to determine the subcellular location of proteins. Computational prediction programs use various machine-learning algorithms that identify sequence features from the primary protein sequence to predict the subcellular location of a protein. These bioinformatic programs have become increasingly important for annotating newly sequenced genes and for providing testable hypotheses regarding protein localization and function (5). However, obviously it is desirable to use experimental data on protein location where this is available. Popular experimental approaches for subcellular determination in Arabidopsis include in vitro protein import studies into isolated organelles, in vivo protein tagging by fluorescent markers and cell fractionation followed by protein detection using enzyme activity measurements, immunolocalization or mass spectrometry (6). Shotgun proteomic studies employing mass spectrometry to identify peptides in purified subcellular compartments result in large, information-rich datasets, whereas targeted fluorescent protein studies allow directed analysis of location and can provide clear evidence of multi-targeting to several locations. Unfortunately, most of these experimental data for Arabidopsis proteins are scattered in the literature and biologists can spend a significant amount of time and effort in searching for all the available localization information. Moreover, a large number of protein localizations can be reported in an article but not listed in the title, abstract or text. Therefore, it is not always easy to access experimental localization data from literature sources. In addition, curated subcellular proteomes and catalogues of GFP targeting information are not readily available as defined datasets.
A number of key databases have been developed to integrate localization data from different sources, such as the Plant Proteomics Database (PPDB) (2), AT_CHLORO (7) and ARAMEMNON (8). ARAMEMNON, e.g., has been designed to overcome the individual limitations of different types of predictors by combining their predictions and including experimental data as further evidence (8). Localization predictions are also reported in PPDB (2) and AT_CHLORO (7) but the assigned subcellular locations are based solely on experimental evidence. Aggregators value-add the use of individual predictors and are recommended when investigating the subcellular location of a protein (9,10).
The SUBcellular localization database for Arabidopsis proteins (SUBA) (4,11) brings together protein localization information for Arabidopsis proteins provided by different prediction algorithms as well as experimental data and annotations. As a central hub for protein localization in Arabidopsis, SUBA has provided access to defined sets of localization data that have been collectively investigated by the research community for the last 15 years. SUBA has been used extensively to define the location of specific proteins in hundreds of reports and also used to assess targeting prediction programs (12,13), identify the localization of protein families (4) and to assess metabolic network models (14,15). By expanding the curated information in SUBA3, including more predictors of targeting, incorporating protein–protein interaction (PPI) data and developing SUBAcon, a Bayesian approach to best estimate a protein’s location in the cell, we have increased the value and reliability of the database.
MATERIALS AND METHODS
Database structure and interface
SUBA3 utilizes the database programming language SQL (Structured Query Language) and is housed on a Linux server running Ubuntu 10.04 LTS. The SUBA3 web browser-based graphical user interface is written in Dynamic Hyper Text Markup Language that makes use of Asynchronous JavaScript and XML (AJAX) to interact with the SUBA server. The back-end of SUBA utilizes a number of PHP scripts that interact with the MySQL tables housing the SUBA data. Making use of complex JavaScript, the interface works best via the Mozilla Firefox, Google Chrome or Safari web browsers but will work on Microsoft Internet Explorer (6 and above). The use of JavaScript allows users to dynamically construct, via the interface, complex Boolean queries without the need to be proficient in SQL. Through the interface, SUBA3 can be easily queried to define subsets of proteins predicted or experimentally found to be located in different parts of the cell. SUBA3 leverages open-source technologies in order to provide a freely available platform at http://suba.plantenergy.uwa.edu.au.
Experimental data sources
The non-redundant nuclear Arabidopsis protein set in SUBA3 was obtained from The Arabidopsis Information Resource (TAIR, release 10) (16). Arabidopsis mitochondrial (117) and chloroplast (87) open reading frame (ORF) sets were obtained from GenBank Y08501 and AP000423, respectively. SUBA3 currently contains a total of 35 388 distinct proteins. Primary attributes for proteins such as molecular weight, average hydropathicity and isoelectric point as well as functional assignments for each Arabidopsis locus were generated as described by Heazlewood et al. (4). Experimental subcellular localizations of proteins by mass spectrometry studies were obtained by searching PubMed (http://www.ncbi.nlm.nih.gov/pubmed) with ‘proteomics’ and ‘Arabidopsis’ or ‘MS’ and ‘Arabidopsis’, whereas localizations of proteins by GFP tagging were obtained using the keyword ‘Arabidopsis’ in combination with ‘fluorescent protein’, ‘GFP’, ‘CFP’, ‘YFP’ or ‘RFP’. Articles were read to determine whether Arabidopsis proteins were localized and the Arabidopsis Genome Initiative (AGI) identifiers with their localizations were extracted directly from the text or from supplementary data. Mass spectrometry-based localizations were obtained from 122 publications and represent 7685 unique proteins. Protein localizations based on GFP tagging studies were obtained from 1074 articles and represent 2477 unique proteins. The textual descriptions were interpreted to fit the 11 subcellular locations defined in SUBA, along with a category of ‘unclear’ for those that could not be fitted to this structure. Additionally, location annotations from literature sources for Arabidopsis proteins add 262 758 entries from TAIR (16), Swiss-Prot (17) and AmiGO (18). PPI datasets of 12 080 protein pairs were obtained by searching the content of the IntAct database for interacting Arabidopsis proteins (19). In addition, 552 interacting PPI pairs were obtained by searching PubMed (http://www.ncbi.nlm.nih.gov/pubmed) using the keywords ‘Arabidopsis’ in combination with ‘interact’, ‘interaction’ or ‘interacting’. The AGI identifiers of interacting Arabidopsis proteins were extracted directly from the text of the articles or from supplementary data.
Subcellular location prediction
Subcellular targeting predictions were carried out using 22 different bioinformatic programs: AdaBoost (20), ATP (21), BaCelLo (22), ChloroP 1.1 (23), EpiLoc (24), iPSORT (25), MitoPred (26), MitoProt (27), MultiLoc2 (28), Nucleo (29), PCLR 0.9 (30), Plant-mPLoc (31), PProwler 1.2 (32), Predotar v1.03 (33), PredSL (34), PTS1 (35), SLPFA (36), SLP-Local (37), SubLoc (38), TargetP 1.1 (5), WoLF PSORT (39) and YLoc (40). Targeting predictions were carried out on the full-length protein sequences obtained from TAIR10 (16).
RESULTS
SUBA curation, interface and the update of experimental data
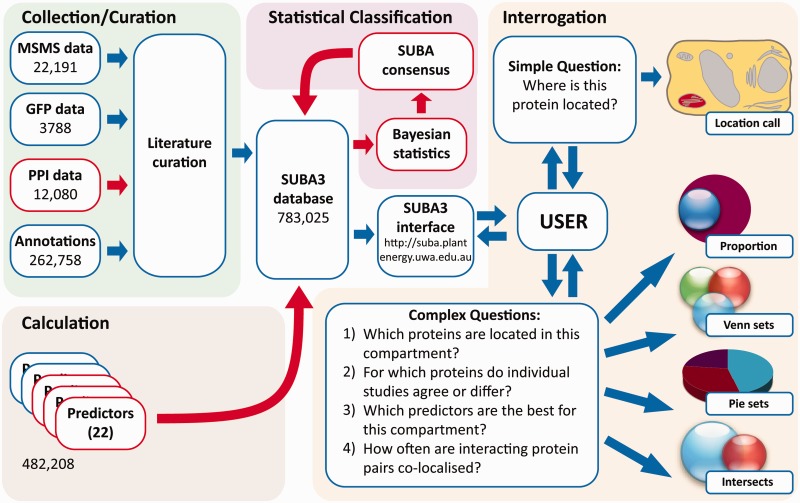
SUBA3 currently comprises 783 025 pieces of subcellular location information for a total of 35 388 non-redundant Arabidopsis proteins (Figure 1). Of these data, 38 059 are calls from experimental evidence curated from the literature as MS/MS, GFP and now PPI data. At the time of writing, there are 22 191 entries based on subcellular proteomic studies, representing 7685 distinct proteins from 122 publications. Additional data from 1074 different publications add 3788 entries based on GFP tagging studies and comprise 2477 distinct proteins (Figure 1). Combined, the experimental data cover a total of 9024 non-redundant proteins localized by mass spectrometry or GFP tagging studies of which 1138 proteins have been localized by both methods. PPI data include 12 080 distinct protein pairs from 534 publications (Figure 1). Further annotation of location from literature sources for Arabidopsis proteins obtained through Swiss-Prot (17) and TAIR (16) contributes a similar number of localizations with 138 393 and 109 340, respectively, whereas AmiGO (18) contributes 15 025 localizations. SUBA3 includes the expansion of the number of predictors from 10 to 22, making use of many new (and better) predictors published in the last 6 years. A total of 482 208 calls are by prediction algorithms. SUBA3 can be queried via a web browser interface, accessible via http://suba.plantenergy.uwa.edu.au (Figure 1). The interface allows users to ask a simple question about one protein or, even with no prior knowledge of SQL, to construct moderately complex SQL queries using drop-down menus and buttons. The interface employs a tabbed design featuring ‘Home’, ‘Search’, ‘Results’ and ‘Help’ tabs.
Figure 1.
SUBA3 curation, calculations, classification and the interface for interrogation. Blue boxes highlight existing sections in SUBA that have been significantly updated, red boxes highlight new sections added in SUBA3.
The primary ‘Search’ tab involves pull-down menus and text boxes for the users’ convenience that can also be used in combination with AND, OR, NOT and parentheses to build complex Boolean queries. Once a query has been submitted, the ‘Results’ page presents a table, which by default contains the AGI identifier, description and localization summary information from predictions, annotations, GFP, mass spectrometry and PPI data. Nearly all retrieved data are linked to a reference in PubMed (http://www.ncbi.nlm.nih.gov/pubmed). Results can be sorted (ascending/descending) by field using the function menu. The function menu is activated by tracking the mouse over the column header and then selecting the emerging arrow. New columns can be added to the ‘Results’ tab window by selecting ‘Columns’ in the function menu and columns can be organized using drag and drop functionality. Thus, users are able to control which data columns are visible and the order in which they are displayed. If further analysis is desired, all results can be downloaded as a tab-delimited file by using the ‘Download All Results’ button. Each AGI identifier in the results page is hyperlinked to a ‘SUBA flatfile’ that provides a variety of information and helpful links. These include detailed subcellular localization information and the capability to include and display GFP images.
Selecting predictors for use for different subcellular compartments
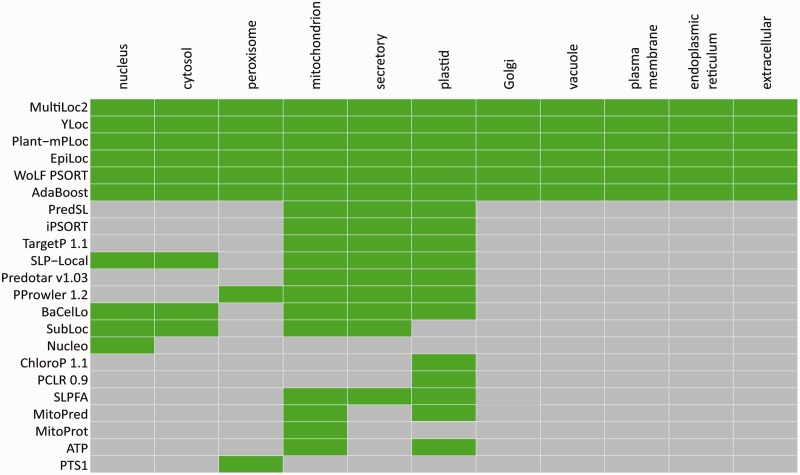
The large increase in number of predictors integrated in SUBA provides an opportunity to analyse their prediction sensitivity and specificity across a range of subcellular locations. A large number of the algorithms that form the basis of these predictors call plastid, mitochondria or the secretory pathway. A smaller number predicts peroxisome and nuclear targeting, and some give null predictions as cytosolic prediction. A different subset provides a breakdown of prediction in the secretory pathway to be vacuole, Golgi, plasma membrane, endoplasmic reticulum and extracellular environment. The coverage of 10 locations defined in SUBA by the various predictors is illustrated in Figure 2.
Figure 2.
Selecting predictors for use for different subcellular compartments. The output of 22 predictors of Arabidopsis protein location across 10 locations are employed in SUBA. The locations predicted by each predictor are shown in green. In total, 6 predictors provide call for all 10 SUBA locations and 16 predictors generate calls for a subset of locations.
Combining experimental data and predictions
Evaluating the large amount of data now available for many Arabidopsis proteins can be difficult for researchers not familiar with the experimental approaches or the prediction software. The limitations of these methods are seldom apparent to non-experts, often leading to overconfidence in the reported results. As more results accumulate, so do conflicting data and predictions, making it increasingly hard to present a clear conclusion for SUBA users. To help reduce this confusion, SUBA now presents a consensus location (SUBAcon) based on Bayesian probabilities calculated from all the experimental data and predictions available for each protein (Figure 1). SUBAcon will be valuable to researchers unsure of how to evaluate the data themselves and also to researchers wishing to automate the evaluation of localization calls for genome-wide analyses (e.g. constructing compartmentalized metabolic networks).
The development of SUBAcon and an assessment of its performance will be described elsewhere; in brief, two Bayesian classifiers have been integrated into SUBA using the 22 subcellular location prediction sets plus the SUBA3-curated GFP and mass spectrometry datasets as inputs into the models. The first classifier evaluates calls to plastid, mitochondrion, peroxisome, cytosol, nucleus and all calls for entry into the secretory pathway; the second classifier treats calls within the secretory pathway to the vacuole, Golgi, plasma membrane, endoplasmic reticulum and to the extracellular environment. Deriving the parameters for the two naive Bayesian models requires estimating the accuracy of the location calls derived from each predictor or experimental approach. This was achieved using a protein ‘reference set’ (RS) compiled by manual analysis of TAIR10 annotation and MapMan (41) evaluation of biochemical pathways and functional groups. Locations in the RS are inferred by function, rather than by localization data alone and the set includes many proteins with dual or multiple locations. This continually improving RS set comprises over 5000 proteins at the time of writing and can be investigated through the SUBA3 search interface using the first row of pull-down menus. To obtain the final probabilities for proteins that enter the secretory pathway, the outputs of the two Bayesian models are combined by multiplying the probability values of locations in the ‘secretory’ model with the probability value of a secretory pathway call from the first model. The probability values of SUBAcon can be viewed by tracking the mouse over the subcellular compartments of the pictographic plant cell heat map on the ‘SUBA3 flatfile’.
PPI data as subcellular location tool
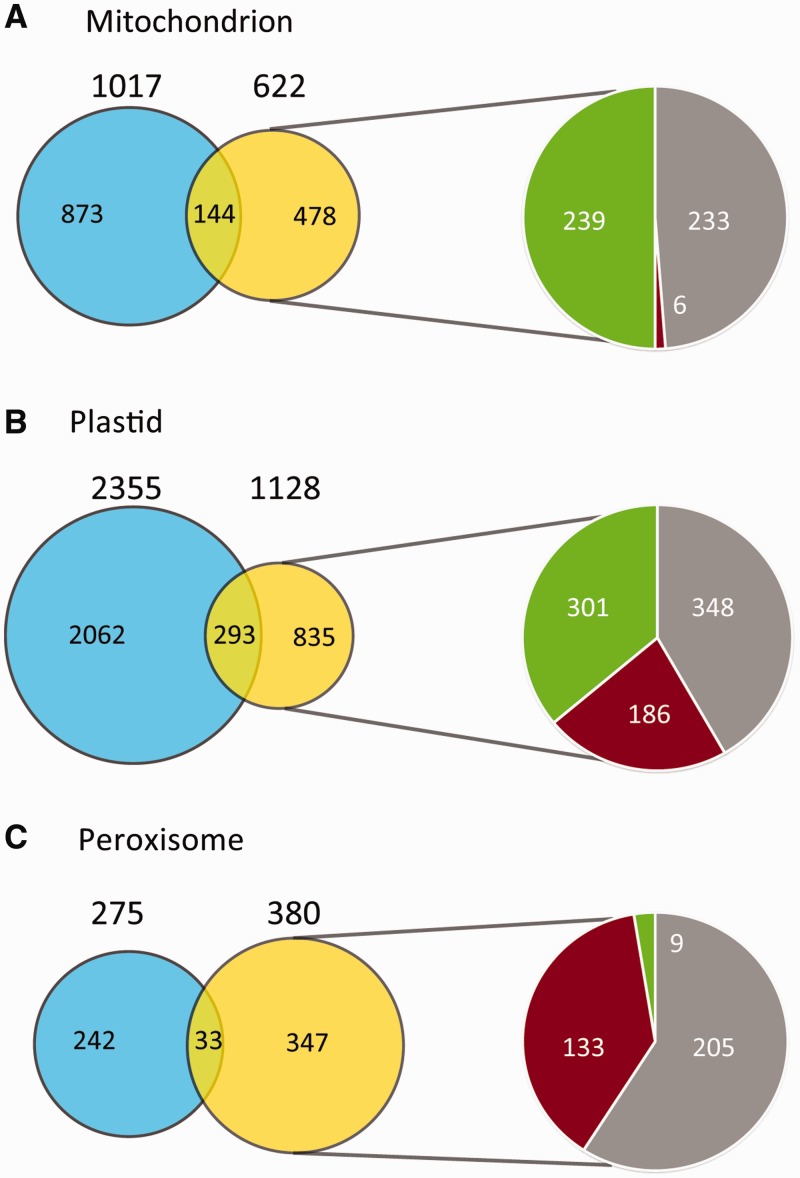
Recently, large experimental PPI datasets for Arabidopsis proteins have been published (42,43), providing a new source of information that can be assessed for its utility to locate proteins within cells. By including these data in SUBA and allowing searches for proteins that are known to interact with a single protein or a subset of search proteins, we are able to use PPI data to extend experimentally defined subcellular proteomes. For example, the mitochondrial experimental proteome of 1017 overlaps with 622 proteins in PPI pairs (Figure 3A), defining 478 proteins that have been shown to interact with a protein experimentally located in mitochondria but which have not been experimentally located in mitochondria themselves. In this set of 478 proteins, 233 have been located elsewhere by MS or GFP, 6 were clearly predicted to be elsewhere, whereas 239 were predicted to be located in mitochondria (Figure 3A). This set of 239 are thus proteins predicted to be mitochondrially located and experimentally interact with proteins known experimentally to be located in mitochondria, making this a strong set of candidates to extend the mitochondrial proteome by ∼20%. Similar analysis of plastids provided a set of 301 proteins (extending the experimental set by ∼15%, Figure 3B), whereas in peroxisomes, this set was only nine proteins (extending the experimental set by ∼3%, Figure 3C). Analysis of these sets of interactions shows that the integration of PPI data can predict binding partners for plastid and mitochondrial heat shock proteins, thioredoxin/glutaredoxins and TPR/PPR proteins and propose unknown function binding partners of peroxin (PEX) proteins in peroxisomes. These PPI datasets of particular compartments can be rapidly generated by any user through the PPI text box below the ‘… protein does/does not interact with proteins(s) in list’ menu row on the SUBA search interface and subsequent analysis of SUBA results in Excel. Once the final set of interacting proteins is obtained, SUBA can be queried again via the PPI text box to obtain matched sets of interacting partners.
Figure 3.
Using PPI data to define extensions of subcellular proteomes. (A) Mitochondria, (B) plastids and (C) peroxisomes. Blue is the experimentally confirmed set by GFP or MSMS, yellow are proteins that interact with the experimental organelle subset, novel interacting proteins (subset of yellow) were analysed for those that were predicted in another compartment (red), predicted in the same compartment (green) or experimentally found in another compartment (grey).
CONCLUSION
Through the combination of wider literature curation, aggregation of predictor calls and integration through the development of SUBAcon, we have significantly extended the richest online aggregation of information on subcellular location of proteins in Arabidopsis. The SUBA3 search interface allows simple inquires about single proteins, as well as very complex queries across these datasets to build subcellular proteomes, compare the performance of different techniques and assess the location of user-defined sets of proteins. Integration of PPI data allows researchers for the first time to easily explore the value of PPI in extending subcellular proteomes of interest. The development of SUBAcon also provides a single probabilistic call of location for all Arabidopsis proteins that will aid system-level studies in Arabidopsis and will continue to improve over time as new experimental data are added to the database.
FUNDING
The Australian Research Council (CE0561495 to A.H.M. and I.S., FT110100242 to A.H.M. and DE120100307 to S.K.T.]; the Government of Western Australia through funding for the WA Centre of Excellence for Computational Systems Biology (DIR WA CoE). Funding for open access charge: The University of Western Australia.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Feldblyum T, Nierman W, Benito MI, Lin XY, et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.Alonso JM, Ecker JR. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat. Rev. Genet. 2006;7:524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- 3.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 4.Heazlewood JL, Tonti-Filippini J, Verboom RE, Millar AH. Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 2005;139:598–609. doi: 10.1104/pp.105.065532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 6.Millar AH, Carrie C, Pogson B, Whelan J. Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell. 2009;21:1625–1631. doi: 10.1105/tpc.109.066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferro M, Brugiere S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell Proteomics. 2010;9:1063–1084. doi: 10.1074/mcp.M900325-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanz SK, Small I. In silico methods for identifying organellar and suborganellar targeting peptides in Arabidopsis chloroplast proteins and for predicting the topology of membrane proteins. Methods Mol. Biol. 2011;774:243–280. doi: 10.1007/978-1-61779-234-2_16. [DOI] [PubMed] [Google Scholar]
- 10.Joshi HJ, Hirsch-Hoffmann M, Baerenfaller K, Gruissem W, Baginsky S, Schmidt R, Schulze WX, Sun Q, van Wijk KJ, Egelhofer V, et al. MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 2011;155:259–270. doi: 10.1104/pp.110.168195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryngajllo M, Childs L, Lohse M, Giorgi FM, Lude A, Selbig J, Usadel B. SLocX: Predicting subcellular localization of Arabidopsis proteins leveraging gene expression data. Front. Plant Sci. 2011;2:43. doi: 10.3389/fpls.2011.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Oliveira Dal'Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 2010;152:579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mintz-Oron S, Meir S, Malitsky S, Ruppin E, Aharoni A, Shlomi T. Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl Acad. Sci. USA. 2012;109:339–344. doi: 10.1073/pnas.1100358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider M, Lane L, Boutet E, Lieberherr D, Tognolli M, Bougueleret L, Bairoch A. The UniProtKB/Swiss-Prot knowledgebase and its Plant Proteome Annotation Program. J. Proteomics. 2009;72:567–573. doi: 10.1016/j.jprot.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu B, Jin YH, Feng KY, Lu WC, Cai YD, Li GZ. Using AdaBoost for the prediction of subcellular location of prokaryotic and eukaryotic proteins. Mol. Divers. 2008;12:41–45. doi: 10.1007/s11030-008-9073-0. [DOI] [PubMed] [Google Scholar]
- 21.Mitschke J, Fuss J, Blum T, Hoglund A, Reski R, Kohlbacher O, Rensing SA. Prediction of dual protein targeting to plant organelles. New Phytol. 2009;183:224–235. doi: 10.1111/j.1469-8137.2009.02832.x. [DOI] [PubMed] [Google Scholar]
- 22.Pierleoni A, Martelli PL, Fariselli P, Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics. 2006;22:e408–e416. doi: 10.1093/bioinformatics/btl222. [DOI] [PubMed] [Google Scholar]
- 23.Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady S, Shatkay H. EpiLoc: a (working) text-based system for predicting protein subcellular location. Pac. Symp. Biocomput. 2008:604–615. [PubMed] [Google Scholar]
- 25.Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 26.Guda C, Guda P, Fahy E, Subramaniam S. MITOPRED: a web server for the prediction of mitochondrial proteins. Nucleic Acids Res. 2004;32:W372–W374. doi: 10.1093/nar/gkh374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 28.Blum T, Briesemeister S, Kohlbacher O. MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 2009;10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkins J, Davis L, Boden M. Predicting nuclear localization. J. Proteome Res. 2007;6:1402–1409. doi: 10.1021/pr060564n. [DOI] [PubMed] [Google Scholar]
- 30.Schein AI, Kissinger JC, Ungar LH. Chloroplast transit peptide prediction: a peek inside the black box. Nucleic Acids Res. 2001;29:E82. doi: 10.1093/nar/29.16.e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One. 2010;5:e11335. doi: 10.1371/journal.pone.0011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawkins J, Boden M. Detecting and sorting targeting peptides with neural networks and support vector machines. J. Bioinform. Comput. Biol. 2006;4:1–18. doi: 10.1142/s0219720006001771. [DOI] [PubMed] [Google Scholar]
- 33.Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 34.Petsalaki EI, Bagos PG, Litou ZI, Hamodrakas SJ. PredSL: a tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics Proteomics Bioinformatics. 2006;4:48–55. doi: 10.1016/S1672-0229(06)60016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 2003;328:581–592. doi: 10.1016/s0022-2836(03)00319-x. [DOI] [PubMed] [Google Scholar]
- 36.Tamura T, Akutsu T. Subcellular location prediction of proteins using support vector machines with alignment of block sequences utilizing amino acid composition. BMC Bioinformatics. 2007;8:466. doi: 10.1186/1471-2105-8-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuda S, Vert JP, Saigo H, Ueda N, Toh H, Akutsu T. A novel representation of protein sequences for prediction of subcellular location using support vector machines. Protein Sci. 2005;14:2804–2813. doi: 10.1110/ps.051597405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hua S, Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics. 2001;17:721–728. doi: 10.1093/bioinformatics/17.8.721. [DOI] [PubMed] [Google Scholar]
- 39.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briesemeister S, Rahnenfuhrer J, Kohlbacher O. YLoc–an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 2010;38:W497–W502. doi: 10.1093/nar/gkq477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 42.Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]