Abstract
Reversible phosphorylation is a key mechanism for regulating protein function. Thus it is of high interest to know which kinase can phosphorylate which proteins. Comprehensive information about phosphorylation sites in Arabidopsis proteins is hosted within the PhosPhAt database (http://phosphat.mpimp-golm.mpg.de). However, our knowledge of the kinases that phosphorylate those sites is dispersed throughout the literature and very difficult to access, particularly for investigators seeking to interpret large scale and high-throughput experiments. Therefore, we aimed to compile information on kinase–substrate interactions and kinase-specific regulatory information and make this available via a new functionality embedded in PhosPhAt. Our approach involved systematic surveying of the literature for regulatory information on the members of the major kinase families in Arabidopsis thaliana, such as CDPKs, MPK(KK)s, AGC kinases and SnRKs, as well as individual kinases from other families. To date, we have researched more than 4450 kinase-related publications, which collectively contain information on about 289 kinases. Users can now query the PhosPhAt database not only for experimental and predicted phosphorylation sites of individual proteins, but also for known substrates for a given kinase or kinase family. Further developments include addition of new phosphorylation sites and visualization of clustered phosphorylation events, known as phosphorylation hotspots.
INTRODUCTION
Regulation of protein activity is a basic necessity for each organism and it is required for functional signaling pathways and metabolism. Activity as well as targeting or interaction with other proteins can be regulated by post-translational modifications. The literature currently describes more than 300 different and substrate-specific modifications of plant proteins (1), underlining the importance of post-translational regulatory mechanisms for studies of plant growth and development as well as their adaptation to stress.
Phosphorylation is one of the most widespread protein modifications and is crucial in signal transduction. Phosphorylation is reversible and catalysed by kinases, which transfer a phosphoryl group from ATP to the hydroxyl group of specific serine, threonine or tyrosine residues within their target proteins (2). In 1987, Hunter postulated that mammalian genomes would encode around 1000 protein kinases (3). Fifteen years later, after sequencing of the human genome, Manning et al. (4) identified 518 kinases (representing <2% of all protein-coding genes in the human genome), which is about half of the initially predicted number of kinases. In contrast, in plants the sequencing of the Arabidopsis genome revealed a remarkable number of around 1000 protein kinase genes, which constitute about 4% of the Arabidopsis genome (2). Additionally, 17 histidine kinases with functionalities resembling the prokaryotic two-component phospho-relay system were identified in the Arabidopsis genome (5).
Plant protein kinases can be grouped in two paraphyletic groups: a larger group of around 600 receptor-like protein kinases (RLKs) and a smaller group of around 400 non-RLK kinases. The biological functions of the majority of RLKs are still poorly understood (6). However, the non-RLK kinases have been relatively well studied, providing information about various aspects of their functions. Some of the kinase families found in plants are clearly related to animal and protozoan kinases, but plant-specific kinases have also been identified. Major groups in plants are the CDPK-SnRK superfamily of protein kinases (7), the three subfamilies of mitogen-activated kinases (8), the AGC kinases (9), the cyclin-dependent protein kinases and the shaggy-like/GSK3 kinases (10).
During the past decade many phosphorylation sites in plant proteins have been identified, especially by mass spectrometry-based large-scale experiments investigating various stress conditions (11–13). In 2007, the PhosPhAt database was launched as a (publicly available?) resource that consolidates our current knowledge of phosphorylation sites identified by mass spectroscopy in the model plant Arabidopsis thaliana (14). In 2010, a new module was introduced into the database, which enables the user to upload any protein sequence of interest and predict potential serine, threonine and tyrosine phosphorylation sites (15). Despite these ongoing improvements to the database, one key aspect of protein phosphorylation was still missing, namely, information about which kinase can phosphorylate which amino acid residue in which proteins.
The literature offers a vast source of information about plant protein kinases—their targets, specificity, regulation, interaction partners and biological functions. Genetic experiments have provided information about the effect of knocking out specific kinases on whole plant physiology and metabolism. However, this information is not readily available in a searchable format, restricting the ability of researchers to make use of this knowledge to interpret their results, especially when dealing with large-scale experiments. Therefore, we aimed to extract information about the substrates, interactions and regulation of plant protein kinases from the literature and make it available as a structured and searchable functionality within the PhosPhAt database.
NEW AND IMPROVED FEATURES OF THE PHOSPHAT DATABASE
A new database module for PhosPhAt was created in which each kinase was paired with its targets, and the nature of each relationship as well as the site of interaction (i.e. phosphorylation sites) was specified. Furthermore, information about the biological context (‘pathway’) of the kinase function and a reference to the original literature was recorded.
Kinase target information
The Arabidopsis kinome consists of around 1000 kinases (16,17), but to date just a small proportion of them has been experimentally characterized and described in literature. We started collecting information on the five major families of non-LRR kinases, namely: (i) calcium-dependent protein kinases (CDPK); (ii) CDPK-related kinases (CDPK-RK); (iii) Snf1-related protein kinases (SnRKs); (iv) cAMP, cGMP and phospholipid-dependent protein kinases (AGCs); and (v) three groups of mitogen-activated kinases (MAPKs). The work will be extended to other kinase families in future.
The CDPK family of plant-specific kinases contains 34 predicted genes/proteins. CDPKs are calcium activated and auto-phosphorylating (7). We found literature information on 22 of the 34 family members, comprising 55 identified target proteins and 10 verified substrate phosphorylation sites. Furthermore, there are eight predicted genes encoding CDPK-related kinases (CDPK-RK), of which at least seven are expressed at the transcript level. CDPK-RKs are angiosperm-specific kinases and apparently have degenerated calcium binding sites, as they show no activation by calcium (7). For two of these kinases, information on two target proteins was retrieved.
The Snf1 protein kinase and related kinases were initially described in yeast, and there are 38 predicted genes belonging to this family in Arabidopsis thaliana. SnRKs are divided into three subgroups: SnRK1, which is homologous to the yeast Snf1 kinase, and the plant-specific subfamilies SnRK2 and SnRK3. Three predicted genes encode SnRK1; two of them (AKIN10 and AKIN11) are expressed and extensively described in literature. We found 19 targets with seven verified phosphorylation sites for SnRK1. Ten predicted genes encode SnRK2s, including the well-studied OST1, and literature information is available for all 10 subfamily members. In total, 66 target proteins of SnRK2 phosphorylation were found, with 25 phosphorylation sites experimentally verified. The SnRK3s, for which there are 25 predicted genes in Arabidopsis, are also known as CBL interacting kinases (CIPKs) and show possible regulation by calcium via the CBL regulatory proteins (7). We found information on 20 SnRK3s in the literature dealing with 31 protein targets and 8 verified substrate phosphorylation sites.
AGC kinases are downstream effectors of intercellular second messengers. Particularly PDK1, a phosphatidylinositol-5-phosphate binding AGC kinase can phosphorylate, and thereby activate, other AGC kinases that lie downstream of it in various signaling pathways. AGC kinases are only found in eukaryotes, and in Arabidopsis there are 39 members of this family, which fall into 6 subgroups (9). Information about 20 AGC kinases was found in the literature, describing 36 protein targets and 23 experimentally verified substrate phosphorylation sites.
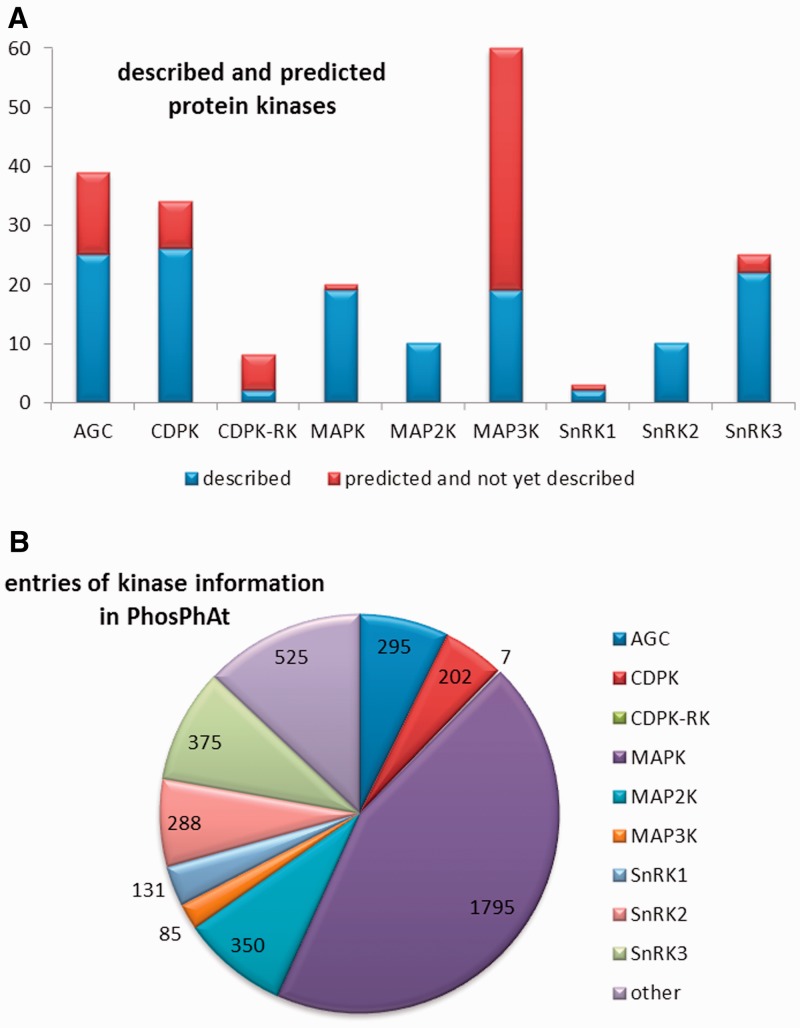
Mitogen-activated protein kinases are eukaryotic kinases that form cascades for intracellular transmission of extracellular stimuli and are classified into three main groups. The MAPK kinase kinases (MAP3Ks) lie furthest upstream and are so-called because they phosphorylate and activate MAPK kinases (MAP2K). In turn, the MAP2Ks phosphorylate MPKs, which are the effector kinases that form the last step in the signaling cascade. Depending on the literature source, 60–80 genes are predicted to encode MAP3Ks in Arabidopsis, and these are clustered in two distinct classes: the STE11-like and the Raf-like kinases, based on the classification of MAP3Ks in mammals. We found published information on 13 of the Arabidopsis MAP3Ks, and 9 out of the 10 MAP2Ks. The MPKs phosphorylate non-kinase target proteins such as enzymes, ribosomal proteins and transcription factors. There are predicted to be 20 MPKs in Arabidopsis and 15 of them are characterized in the literature (8). For the MAPK family as a whole, a total of 137 target proteins and 21 verified substrate phosphorylation sites were retrieved from literature. Information on an additional 562 possible phosphorylation targets for MAPKs, derived from peptide array experiments (18), was also incorporated into PhosPhAt. An overview of the numbers of kinases for which information was found and the distribution of this information between the major kinase families is presented in Figure 1.
Figure 1.
Overview of the kinases within the non-RLK families that have been researched so far. (A) Number of genes encoded in the Arabidopsis genome and the number of kinases described in literature up to date. (B) Representation of regulatory information about the different kinase families as represented by the numbers or regulatory interactions entered to the database.
In addition to the five major kinase families, we also systematically collected information about cysteine-rich receptor-like kinases (CRK) and other receptor-like kinases, along with kinases involved in brassinosteroid signaling. Some information on Shaggy-like/GSK3 kinases was also extracted from the literature, but in a less systematic way. All of these data are collected under the heading ‘other kinases’ in Figure 1. In addition to data-mining the literature, we also extracted information on kinase target protein–protein interactions from the TAIR database (www.arabidopsis.org), kinase target-phosphorylation data from the UniProt database (www.uniprot.org). The information available from these two databases extends also to kinase families that we have not yet curated in detail.
Classification of kinase–target relationships
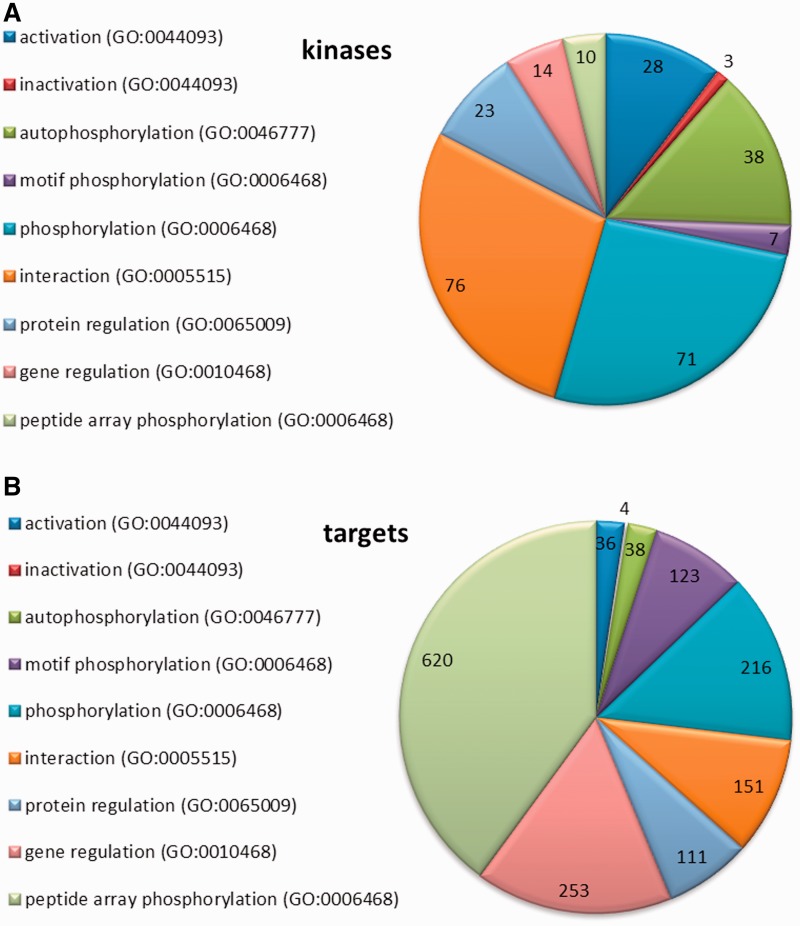
We used standardized vocabularies for the classification of kinases, following the principles employed for Gene Ontology (GO) and Molecular Interaction (MI) terms (19–21). Four major types of kinase–target relationships were defined: (i) ‘regulation’ (including regulation at either the gene or protein level); (ii) ‘activation’ and ‘inactivation’; (iii) ‘interaction’; and (iv) ‘phosphorylation’ (see also ‘Kinome’ tab on the PhosPhAt main website). A representation of the distribution of these different relationships is shown in Figure 2 for the kinases (Figure 2A) and the targets (Figure 2B).
Figure 2.
Representation of the kinase–target relationships entered to the database. (A) Regulatory information collected for kinases (B) Regulatory information entered for kinase targets.
The least-specific category is ‘regulation’, which refers to any relationship in which a kinase affects the expression of another protein at either the gene or protein level. Genes or proteins regulated by particular kinases were entered in our database as kinase regulation targets. We define ‘gene regulation’ as a direct or indirect regulation of genes by a kinase according to GO:0010468; regulation of gene expression. For the most part, these relationships have been experimentally characterized by gene expression analysis in kinase knock-out or overexpression lines (22). The sub-category ‘Protein regulation’ is used when there is an experimentally verified relationship between the kinase and the target protein itself, but further information about the nature of the relationship or the involvement of other proteins is missing (GO:0065009; regulation of molecular function). Such information on protein regulation was usually derived from experiments with mutant lines or stress treatments. For example, a knock-out of the AGC kinase PINOID (At2g34650) induces an apical-to-basal shift in the intracellular localization of the PIN auxin efflux proteins (At1g73590; At5g57090) (23), which disturbs the auxin polar transport and leads to defective organogenesis. However, this study did not resolve whether there is a direct relationship between PINOID and PIN (e.g. protein–protein interaction or phosphorylation) or if are any other proteins involved in the signal transduction that could be regulated by PINOID. Thus, in the absence of more specific information, we would classify this relationship as a ‘regulation’ of PIN by PINOID. However, in this particular example, later experiments showed that PINOID does indeed phosphorylate PIN1 and PIN2 (24–27).
Relationships between kinases and their targets were assigned to the second category—‘activation/inactivation’—when there was experimental evidence of activation (corresponding to GO:0044093; positive regulation of molecular function) or inactivation (GO:0044092; negative regulation of molecular function) of the target at a biochemical level, but the precise nature of the regulation was not defined in the respective publication. Examples in this category include kinases that are targets of another kinase (like in the MAPK cascade) or where the activation/inactivation of a kinase occurs via a receptor protein, a phosphatase or some other regulatory factor. Within this category, the mechanism underlying the activation/inactivation of the target protein is not defined, but could include any of the following: phosphorylation, conformational change, binding of another regulatory factor or some other form of direct interaction between the kinase and its target protein. Experimentally, evidence of activation or inactivation came from transient expression of wild type and mutated kinase proteins in protoplasts, or in vitro activity assays with native or artificial target proteins.
Transient expression studies in protoplasts demonstrated a cascade of activation/inactivation connections for the MAPK pathway (28). It was shown that after ethylene perception, CTR1 (At5g03730; a MAP3K) activates MKK9 (At1g73500; a MAP2K) (28). In turn, MKK9 activates the downstream MAP kinases MPK3 (At3g45640) and MPK6 (At2g43790). However, from that work it was not clear from the reported experiments if the activation of MKK9, or MPK3 and MPK6 was due to phosphorylation. We therefore classified this relationship as an ‘activation’ of a kinase (MAP2K) by an upstream kinase (MAP3K). In another later publication, it was shown that MKK9 can indeed ‘phosphorylate’ MPK6 (29), and we entered this information as a separate record to the database. In another example, experiments with the AGC kinase PDK1 (At5g04510) demonstrated that this kinase needs auto-phosphorylation for full activity (30). Another AGC kinase, S6K2 (At3g08720), is a phosphorylation target of PDK1, and their interaction can be modulated by 14-3-3 proteins. In vitro assays containing PDK1, S6K2 and individual 14-3-3 protein isoforms revealed that 9 out of the 12 tested 14-3-3 proteins enhanced PDK1 activity and 1 suppressed its activity, but the mechanism remains unknown. We therefore classified these relationships as activation/inactivation of PDK1 by regulatory factors.
The third type of connection between kinases and their targets is by a direct ‘interaction’ (GO:0005515; protein binding and MI:0914; association). Protein–protein interactions, experimentally proven by classic methods like yeast-two-hybrid, pull-down assays or fluorescence overlay experiments, are often accompanied by phosphorylation of a target by the kinase, but can also have a regulatory function without covalent modification. There are many kinase interactions described in literature, but phosphorylation of the target protein been demonstrated in relatively few cases, and for the most part the nature and function of the kinase interaction remains unresolved.
The most specific category of biological relationship between a kinase and a target protein is ‘phosphorylation’, defined as the addition of a phosphate group to an amino acid residue in the target protein (GO:0006468; protein phosphorylation and MI:0217; phosphorylation reaction). Our definition of phosphorylation also includes phosphorylation of a kinase by itself (autophosphorylation, GO:0046777; protein autophosphorylation). Additionally, we also recorded information on ‘dephosphorylation’, defined as the removal of a phosphate group by a phosphatase (GO:0006470; protein dephosphorylation and MI:0203; dephosphorylation reaction). Experimentally, phosphorylation was demonstrated by in vitro phosphorylation assays, or by peptide mass spectrometry after a biological treatment of a kinase knockout mutant or a kinase overexpression line. Another approach for identifying phosphorylation partners of specific kinases was through the analysis of phosphorylation sequence motifs (motif phosphorylation) (31). Screening of a semi-degenerate peptide array, in conjunction with in silico analysis of the phosphorylated peptide sequences, has been used to define consensus target site motifs for four SnRK kinases (At3g01090; At1g60940; At5g35410; At4g33950), one CDPK (At3g57530) and the PP2C phosphatase HAB1 (At1g72770). With this information, it was possible to search the Arabidopsis proteome database to identify putative target proteins for these kinases and the HAB1 phosphatase. In addition, results from in vitro phosphorylation experiments using purified kinases on protein or peptide arrays are also included in the PhosPhAt database (32–35).
Pathway: biological context of kinase function
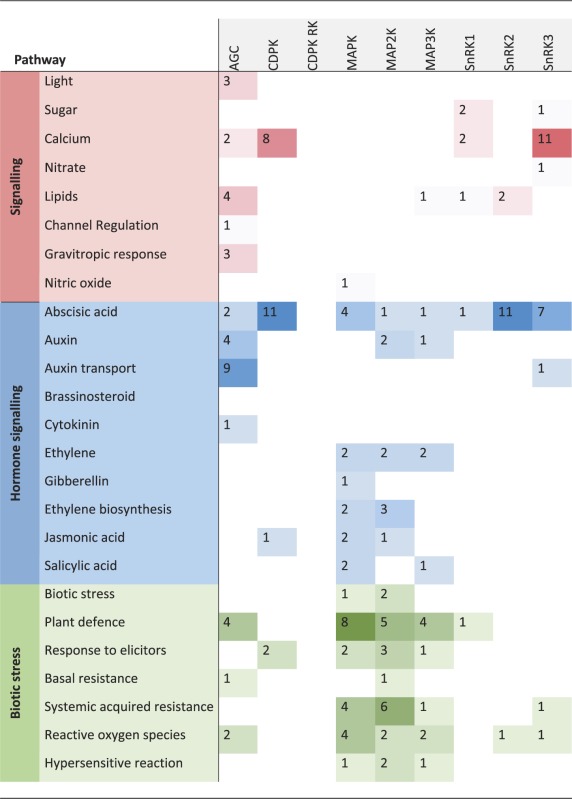
Kinases are involved in many different signaling and regulatory pathways. Some kinases can be quite promiscuous, having multiple targets but many, perhaps most, have very specific targets and functions. Where information on the function of a kinase is available, we assigned it to a ‘pathway’, based on the biological process (GO:0008150; biological process and MI:0619; pathway) it is involved in. For 33 kinases the literature-provided information about their respective pathways, but nothing about their potential targets.
Phenotypic analyses of kinase knockout mutants or overexpression lines, and knowledge of their regulatory or interaction partners, provide the most obvious sources of information about the biological functions of kinases. Such information is often already captured within the annotation of the kinase itself, or its assignment to a pathway or functional category within the MAPMAN (36) ontology, or comparable functional classifications. The ‘pathway’ assignments for kinases in the PhosPhAt database are based on published experimental data, such as a visible phenotype of a mutant line, or evidence of interactions or phosphorylation of target proteins that have a known function. The most common assignment was to hormone signaling pathways, of which 50% were attributed to abscisic acid signaling. Other highly represented pathways were in biotic and abiotic stress, and in development (Table 1). Other pathway functions (not shown in Table 1) included cell division and proliferation, carbon, phosphate and nitrate metabolism, protein-related pathways and transport.
Table 1.
Overview of the most represented ‘pathways’ the systematically researches kinase families are involved in Counts correspond to number of kinases within a given family, which are involved in the specific biological context
 |
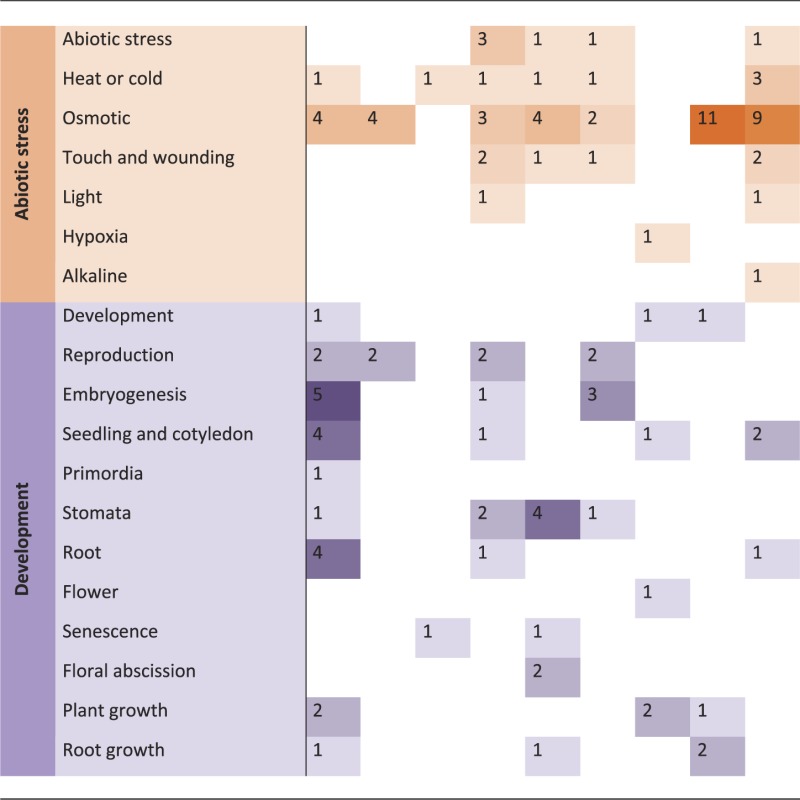 |
Based on MAPMAN functional categorizations (37), we defined ‘signaling’ as the involvement of a kinase in a signal transduction pathway initiated by a stimulus, with sub-classifications based on the nature of that stimulus: ‘light’, ‘sugar’, ‘calcium’, ‘lipids’ (myo-inositol and phosphatidic acid), ‘channel regulation’ and ‘gravitropic response’. Signal transduction involving hormones included all upstream signals leading to hormone biosynthesis as well as downstream signals following hormone perception. The general term ‘hormone signaling’ was further qualified with ‘biosynthesis’ whenever this was clearly specified in the literature or was obvious from target information. We found kinases involved in abscisic acid-regulated pathways, auxin and auxin transport, brassinosteroid, cytokinin, ethylene, gibberellin, jasmonic acid and salicylic acid signaling. The second largest category of pathways was biotic and abiotic stress, where there is evidence that a kinase is involved in signal transduction after stress application. We divided biotic stress into more detailed sub-categories, such as ‘plant defense’, ‘response to elicitors’, ‘basal and systemic acquired resistance’, ‘reactive oxygen species’ and ‘hypersensitive reaction’. Likewise, ‘abiotic stress’ was sub-classified into responses to ‘heat or cold stress’, ‘osmotic stress’ (including drought and salt stress), ‘touch and wounding’, ‘light’, ‘alkaline stress’ and ‘hypoxia’. The third most common category, ‘development’, includes kinases shown to be involved in organ development and patterning, or more generally in regulation of growth and biomass production. The ‘development’ category was subdivided into reproduction and embryogenesis, seedling/cotyledon, primordia, stomata, root or flower development, senescence, floral abscission, root growth or plant growth in general, based on commonly used themes within the developmental field.
Implementation in the PhosPhAt database
The information about kinase targets can be searched via the ‘kinase targets’ tab in the left column by entering one or multiple Arabidopsis gene identifiers. The search results are displayed in a table format, with the query kinase in the ‘kinase’ column and the respective targets listed in the ‘target’ column. The type of relationship is specified for each target, and GO- or MI-terms describing this relationship can be visualized via a context menu in the column head. The reference to the original experimental evidence is provided through the PubMed ID number (http://www.ncbi.nlm.nih.gov/pubmed/).
By checking the selection box for one of the kinase–target relationships, the user can then select the ‘Prediction’ tab to obtain a sequence view of either the kinase or its target. The protein sequence is annotated to show both predicted (highlighted in green) and experimentally determined (in bold) phosphorylation sites. Where the kinase that phosphorylates a particular residue is known, the residue is highlighted in bold orange. Phosphorylation hotspots (38) are highlighted in light green and Pfam (http://pfam.sanger.ac.uk) domains in yellow. Positioning the mouse cursor on any of the highlighted phosphorylation sites brings up additional information below the sequence, such as details of the experimentally identified peptide sequence, prediction scores or the phosphorylating kinase.
An advanced query option allows the user to search for kinase target combinations, with the search constrained to particular pathways or interaction types. Furthermore, kinase target information can be retrieved for whole kinase families through the ‘kinase family’ tab. Kinase target pairs in the search results page can be exported as a tab delimited text file by selecting the export option in the top-right corner. The user can choose whether to export all of the data or just the selected items from the list.
In general, information in PhosPhAt can also be queried through the MASCP Gator portal (http://gator.masc-proteomics.org/) (39) and therefore in principle can also be made part of the proposed Arabidopsis Information Portal (40,41).
Other updates to the PhosPhAt resource
The integration of searchable information about plant protein kinases represents a major extension of the PhosPhAt database, providing a valuable new resource for the science community. Some other minor modifications and additions have been made to further improve the functionality and scope of the database.
Motif search
The motif search function was already implemented in PhosPhAt (15), but has now been made more directly accessible by introducing a separate search tab in the left-hand column. Furthermore, the new implementation allows users to input their own custom motif sequence(s) when searching the database.
Phosphorylation hotspots
Phosphorylation hotspots are regions in a protein with clustered phosphorylation sites (38). These hotspots may have particular functions in integration of signals from different pathways and in redirecting proteins to a new intracellular location. Experimentally determined as well as predicted phosphorylation hotspots (42) are now hosted in the PhosPhAt database and are highlighted in the protein sequence view.
General update of database content
The database content of plant phosphorylation sites has been updated with information from recent large-scale phosphoproteomic studies (11–13,43,44).
CONCLUSION
The PhosPhAt database was initially established as a searchable repository of information about experimentally proven phosphorylation sites in plant proteins, with later developments allowing in silico prediction of phosphorylation sites. We have now greatly extended the scope of the database by integrating curated information about plant protein kinases, which has been extracted from over 4000 original publications and various databases containing genomic and proteomic data from Arabidopsis.
Following this major expansion, the user is now able to search the PhosPhAt database from two viewpoints. The first, and original, type of query is focused on knowing whether a protein of interest to the user is a target for phosphorylation. Known and predicted phosphorylation sites are displayed within the putative target protein sequence, and the search results now include any available information about the phosphorylating kinase(s). The second, and novel, type of query takes the protein kinase itself as the starting point, and provides the user with information about its targets, interaction partners, biological function and regulation, along with links to the corresponding literature sources. Considerable progress has been made in understanding brassinosteroid signaling (45), MAP kinase cascades (8) and the role of CDPKs and SnRKs in ABA signaling (46), and information about the kinases involved in these pathways is collated in published reviews. However, our knowledge about the role of phosphorylation in most other signaling pathways in plant cells is fragmentary and highly dispersed, making it difficult to access from the literature. We have now compiled data for almost 300 of the predicted 1000 protein kinases in Arabidopsis from the literature and other sources, and made this information more readily available in an integrated and searchable format within the PhosPhAt database. In the future we aim to systematically extend the database to increases information about kinase families that are so far under-represented, and update existing entries as new information becomes available. We present the updated and extended PhosPhAt database as a freely available resource in the hope that it will be a valuable contribution to the scientific community, particularly in data-mining efforts and interpretation of large-scale systems biology experiments.
FUNDING
Funding for open access charge: Max Planck Gesellschaft, Germany.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank John Lunn for his valuable comments on the manuscript text.
REFERENCES
- 1.Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Champion A, Kreis M, Mockaitis K, Picaud A, Henry Y. Arabidopsis kinome: after the casting. Funct. Integr. Genom. 2004;4:163–187. doi: 10.1007/s10142-003-0096-4. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- 4.Manning G, Whyte DB, Martinez R, Hunter R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1914. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 5.Grefen C, Harter K. Plant two-component systems: principles, functions, complexity and cross talk. Planta. 2004;219:733–742. doi: 10.1007/s00425-004-1316-4. [DOI] [PubMed] [Google Scholar]
- 6.Shiu S-H, Bleecker AB. Receptor-like kinases form Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, et al. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 9.Bögre L, Ökresz L, Henriques R, Anthony RG. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003;8:424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- 10.Jonak C, Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 2002;7:457–461. doi: 10.1016/s1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- 11.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl Acad. Sci. USA. 2010;107:15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010;153:1161–1174. doi: 10.1104/pp.110.157347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelsberger WR, Schulze WX. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen starved Arabidopsis seedlings. Plant J. 2012;69:978–995. doi: 10.1111/j.1365-313X.2011.04848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008;36:D1015–D1021. doi: 10.1093/nar/gkm812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38:D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gribskov M, Fana F, Harper J, Hope DA, Harmon AC, Smith DW, Tax FE, Zhang G. PlantsP: a functional genomics database for plant phosphorylation. Nucleic Acids Res. 2001;29:111–113. doi: 10.1093/nar/29.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tchieu JH, Fana F, Fink JL, Harper J, Nair TM, Niedner RH, Smith DW, Steube K, Tam TM, Veretnik S, et al. The PlantsP and PlantsT Functional Genomics Databases. Nucleic Acids Res. 2003;31:342–344. doi: 10.1093/nar/gkg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009;23:80–92. doi: 10.1101/gad.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cote RG, Jones P, Apweiler R, Hermjakob H. The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinform. 2006;7:1–7. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fragoso S, Espíndola L, Páez-Valencia J, Gamboa A, Camacho Y, Martínez-Barajas E, Coello P. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol. 2009;149:1906–1916. doi: 10.1104/pp.108.133298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 24.Dhonukshe P, Huang F, Galvan-Ampudia CS, Mähönen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137:3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- 25.Huang F, Zago MK, Abas L, van Marion A, Galvan-Ampudia CS, Offringa R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell. 2010;22:1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 27.Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 2009;150:722–735. doi: 10.1104/pp.108.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signaling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou C, Cai Z, Guo Y, Gan S. An arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009;150:167–177. doi: 10.1104/pp.108.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otterhag L, Gustavsson N, Alsterfjord M, Pical C, Lehrach H, Gobom J, Sommarin M. Arabidopsis PDK1: identification of sites important for activity and downstream phosphorylation of S6 kinase. Biochimie. 2006;88:11–21. doi: 10.1016/j.biochi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nature Biotech. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 32.Feilner T, Hultschig C, Lee JM, Meyer S, Immink RGH, Koenig A, Possling A, Seitz H, Beveridge A, Scheel D, et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics. 2005;4:1558–1568. doi: 10.1074/mcp.M500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, Leung J, Merlot S. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One. 2010;5:e13935. doi: 10.1371/journal.pone.0013935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlad F, Droillard MJ, Valot B, Khafif M, Rodrigues A, Brault M, Zivy M, Rodriguez PL, Merlot S, Laurière C. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 2010;63:778–790. doi: 10.1111/j.1365-313X.2010.04281.x. [DOI] [PubMed] [Google Scholar]
- 35.Vlad F, Turk BE, Peynot P, Leung J, Merlot S. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 2008;55:104–117. doi: 10.1111/j.1365-313X.2008.03488.x. [DOI] [PubMed] [Google Scholar]
- 36.Usadel B, Nagel A, Thimm O, Redestig H, Bläsing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 38.Riano-Pachon DM, Kleesen S, Neigenfind J, Durek P, Weber E, Engelsberger WR, Walther D, Selbig J, Schulze WX, Kersten B. Proteome-wide survey of phosphorylation patterns affected by nuclear DNA polymorphisms in Arabidopsis thaliana. BMC Genomics. 2010;11:411. doi: 10.1186/1471-2164-11-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joshi HJ, Hirsch-Hoffmann M, Bärenfaller K, Gruissem W, Baginsky S, Schmidt R, Schulze WX, Sun Q, van Wijk KJ, Egelhofer V, et al. MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 2011;155:259–270. doi: 10.1104/pp.110.168195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Arabidopsis Informatics Consortium,T. An international bioinformatics infrastructure to underpin the Arabidopsis community. Plant Cell. 2010;22:2530–2536. doi: 10.1105/tpc.110.078519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.International Arabidopsis Informatics Consortium,T. Taking the next step: building an Arabidopsis Information Portal. Plant Cell. 2012;24:2248–2256. doi: 10.1105/tpc.112.100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christian JO, Braginets R, Schulze WX, Walther D. Characterization and prediction of protein phosphorylation hotspots in Arabidopsis thaliana. Front. Plant Sci. 2012;3:207. doi: 10.3389/fpls.2012.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Höhenwarter W, Weckwerth W. Comparative analysis of phytohormone – responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and MAPA. Plant J. 2010;63:1–17. doi: 10.1111/j.1365-313X.2010.04218.x. [DOI] [PubMed] [Google Scholar]
- 44.Mayank P, Grossman J, Wuest S, Boisson-Dernier A, Roschitzki B, Nanni P, Nühse T, Grossniklaus U. Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J. 2012;72:89–101. doi: 10.1111/j.1365-313X.2012.05061.x. [DOI] [PubMed] [Google Scholar]
- 45.Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingam A, Wang ZY. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asano T, Hayashi N, Kikuchi S, Ohsugi R. CDPK mediated abiotic stress signaling. Plant Signal Behav. 2012;7:817–821. doi: 10.4161/psb.20351. [DOI] [PMC free article] [PubMed] [Google Scholar]