Abstract
In this work, we developed a family-based database of UUCD (http://uucd.biocuckoo.org) for ubiquitin and ubiquitin-like conjugation, which is one of the most important post-translational modifications responsible for regulating a variety of cellular processes, through a similar E1 (ubiquitin-activating enzyme)–E2 (ubiquitin-conjugating enzyme)–E3 (ubiquitin-protein ligase) enzyme thioester cascade. Although extensive experimental efforts have been taken, an integrative data resource is still not available. From the scientific literature, 26 E1s, 105 E2s, 1003 E3s and 148 deubiquitination enzymes (DUBs) were collected and classified into 1, 3, 19 and 7 families, respectively. To computationally characterize potential enzymes in eukaryotes, we constructed 1, 1, 15 and 6 hidden Markov model (HMM) profiles for E1s, E2s, E3s and DUBs at the family level, separately. Moreover, the ortholog searches were conducted for E3 and DUB families without HMM profiles. Then the UUCD database was developed with 738 E1s, 2937 E2s, 46 631 E3s and 6647 DUBs of 70 eukaryotic species. The detailed annotations and classifications were also provided. The online service of UUCD was implemented in PHP + MySQL + JavaScript + Perl.
INTRODUCTION
The 2004 Nobel Prize in Chemistry was awarded to Aaron Ciechanover, Avram Hershko and Irwin Rose for their seminal discovery of the ubiquitin conjugation that targets proteins for degradation in an ATP-dependent manner (1–4). Previously, ubiquitin was isolated as a heat-stable protein with 76 aa (3), while further analyses revealed that the ‘ubiquitin kiss’ functions as a molecular death-tag through the ubiquitination, which reversibly and covalently forms an isopeptide bond between the C-terminal (–GG) carboxyl group of a ubiquitin protein and the ε-amino group of lysine residues or, less commonly, other types of residues of a substrate protein (4,5). The protein substrates can be modified by conjugating with mono- or polyubiquitins (4). Beyond degradation, non-proteolytic functions of ubiquitin conjugation were also demonstrated (6,7). For example, the K63-linked ubiquitin chain is implicated in DNA repair and endocytosis (7), whereas monoubiquitination plays an important role in histone regulation, virus budding and DNA repair (6,7).
The enzymatic process of the ubiquitin conjugation is a sequential three-step cascade (4). First, the C-terminal glycine (G) of ubiquitin is activated by a ubiquitin-activating enzyme (E1) to form an E1–Ub thioester in an ATP-dependent manner. Then the activated ubiquitin is transferred to the active site cysteine (C) residue of a ubiquitin-conjugating enzyme (E2). Finally, a ubiquitin-protein ligase (E3) removes the ubiquitin from the E2 and forms a covalent link between ubiquitin and a protein substrate (4). Also, ubiquitination is reversible by numerous deubiquitination enzymes (DUBs) (8). Analogous to ubiquitin, other components of superfamily of ubiquitin-like modifiers, such as SUMO, ISG15, Rub1/Nedd8 and Atg8/12 are also adopted the similar catalytic procedures (9). The ubiquitin and ubiquitin-like conjugation regulate a large number of cellular processes, such as cell cycle, signal transduction, apoptosis and autophagy (4,9), while aberrant modification is implicated in numerous pathologies, such as neurodegenerative disorders, inflammatory diseases and cancers (7,10). In this regard, identification of E1s, E2s, E3s and DUBs is fundamental for understanding the regulatory roles of ubiquitin and ubiquitin-like conjugation and provides candidate drug targets for further biomedical consideration (11).
After the first E1 ligase was isolated in 1981 (12), identification and classification of E1s, E2s, E3s and DUBs had emerged to be a great challenge. In 2003, with the completion of RIKEN FANTOM-2 project, the total ubiquitin-associated enzymes were identified as 835, 764, 320, 785, 162 and 145 for Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Schizosaccharomyces pombe and Saccharomyces cerevisiae, respectively (13). In particular, there were 16 E1s, 53 E2s, 527 E3s and 74 DUBs characterized in H. sapiens and roughly classified by InterPro domains (13). Later, the number of human putative DUBs was refined as 95 and further classification was based on phylogenetic relations (8). In 2003, the first public database of PlantsUBQ was constructed with E1s, E2s and E3s in Arabidopsis (14). Later, the SCUD database contained 11 E2s, 42 E3s, 20 DUBs and 940 substrates in S. cerevisiae (15). In 2009, the plantsUPS was developed with E1s, E2s and E3s in seven plants (16). Recently, the hUbiquitome database collected 1 E1, 12 E2s, 138 E3s, 17 DUBs and 279 substrates of H. sapiens from the scientific literature (17). Although a number of studies were performed, an integrative data resource is still not available.
In this work, we manually collected 26 E1s, 105 E2s, 1003 E3s and 148 DUBs for ubiquitin and ubiquitin-like conjugation systems from the scientific literature (Table 1). Based on previously established rationales (8,9,18–23), we classified E1s, E2s and DUBs into one, three and seven families, respectively. Also, we classified E3s into a hierarchical structure with five levels, including class, group, subgroup, family and single E3. Totally, we obtained 2 classes, 7 groups, 4 subgroups and 19 families for E3s. With HMMER (24), we totally constructed 1, 1, 15 and 6 hidden Markov model (HMM) profiles for E1s, E2s, E3s and DUBs at the family level, respectively. Then we used the HMM profiles to computationally characterize 738 E1s, 2797 E2s, 43 881 E3s and 6516 DUBs from 70 eukaryotic organisms, respectively. For families without HMM profiles, we additionally conducted an ortholog search to detect 140 E2s, 2750 E3s and 131 DUBs. The classification information was provided, while the detailed annotations from Ensembl (25) and UniProt (26) databases were also integrated. Finally, a comprehensive database of ubiquitin and ubiquitin-like conjugation (UUCD) was developed with 56 949 enzymes for ubiquitin and ubiquitin-like conjugation across 70 eukaryotic species.
Table 1.
The data statistics of known E1, E2, E3 and DUB proteins
| Organism | E1 | E2 | E3 | DUB | Total |
|---|---|---|---|---|---|
| Homo sapiens | 8 | 39 | 475 | 91 | 613 |
| Mus musculus | 2 | 6 | 71 | 14 | 93 |
| Drosophila melanogaster | 0 | 5 | 46 | 4 | 55 |
| Caenorhabditis elegans | 5 | 6 | 57 | 2 | 70 |
| Saccharomyces cerevisiae | 3 | 16 | 78 | 19 | 116 |
| Schizosaccharomyces pombe | 2 | 4 | 39 | 6 | 51 |
| Arabidopsis thaliana | 5 | 27 | 182 | 10 | 224 |
| Others | 1 | 2 | 55 | 2 | 60 |
| Total | 26 | 105 | 1003 | 148 | 1282 |
From the scientific literature, we manually collected experimentally identified E1s, E2s, E3s and DUBs, respectively.
CONSTRUCTION AND CONTENT
Data collection and curation
Although there are up to 17 types of ubiquitin-like proteins that can covalently modify other molecules, only nine of them specifically recognize protein substrates (9). Thus, we searched the PubMed with multiple keywords, including ‘ubiquitin’, ‘ubiquitination’, ‘ubiquitylation’, ‘SUMO’, ‘sumoylation’, ‘NEDD8’, ‘neddylation’, ‘Atg8’, ‘Apg8’, ‘Atg12’, ‘Apg12’, ‘Urm1’, ‘ISG15’, ‘isgylation’, ‘UFM1’ and ‘FAT10’, respectively. Totally, we collected 26 E1s, 105 E2s, 1003 E3s and 148 DUBs for ubiquitin and ubiquitin-like conjugation in eukaryotes (Table 1 and Supplementary Table S1). From Ensembl (25) and UniProt (26) databases, the full-length sequences of the enzymes were obtained. The functional domain information was taken from the annotations in the UniProt database and further verified by searching the Pfam database (27). Moreover, the complete proteome sequences were downloaded for 70 eukaryotes, including 54 animals, 14 plants and 2 fungi, from Ensembl (release version 64, http://www.ensembl.org/), EnsemblPlants (release version 11, http://plants.ensembl.org/) and EnsemblFungi (release version 11, http://fungi.ensembl.org/), respectively (25).
The Classification
As previously described, the known E1s can be distinguished from other proteins by the occurrence of ThiF/MoeB domains (9). Thus, all E1s were classified into a single family. Also, the E2s were categorized into three families based on their functional domains, including UBC (ubiquitin-conjugating), UEV (ubiquitin-enzyme variant) and Other (unclassified) (18). Moreover, DUBs for ubiquitin were classified into five families, including UCH (ubiquitin C-terminal hydrolase), USP (ubiquitin-specific protease), OTU (ovarian tumor), Josephin (Machado–Joseph disease, MJD) and JAMM (JAB1/MPN/Mov34 metalloenzyme) (8). In addition, the DUBs for ubiquitin-like conjugations, such as SENPs for SUMO, were classified into the family of ULP (ubiquitin-like protease). The Other family contained unclassified DUBs (8).
The classification of E3 ligases is complicated, because a considerable number of proteins participate in forming E3 ligase complexes as adaptors without E3 activities (19–23). In this regard, we first classified E3-associated proteins into two classes as E3 activity and E3 adaptor (19–22). Enzymes in the E3 activity class were further categorized into four groups and seven families, including HECT (with one family of HECT), RING (two families as RING and U-box), N-recognin (three families as UBR-box, N-domain and Other) and Other (with one family of Other). Proteins in the E3 adaptor class were classified into 2 groups and 12 families, including Cullin RING (10 families as Cullin, F-box, SKP1, SOCS/VHL/BC-box, Elon-B/C, BTB_3-box, BTB_Other, DWD, DDB1 and Other) and APC/C (two families as CDC20 and APC/C). As different Cullins determine the specific compositions of distinct Cullin RING E3 ligase complexes (21,23), we further classified adaptors in the Cullin RING group into five subgroups, including SCF (Cullin 1 and 7), ECS (Cullin 2 and 5), BCR (Cullin 3), DCX (Cullin 4A/B) and Other (with one family of Other).
Proteome-wide identification
As the number of proteins is limited for several families, we totally constructed 1, 1, 15 and 6 HMM profiles for E1, E2, E3 and DUB families, respectively. The functional domain sequences for each family were firstly aligned by MUSCLE (http://www.drive5.com/muscle/, version 3.8.31), a widely used program for multiple sequence alignment (28). Then we used the hmmbuild program in the HMMER 3.0 package (http://hmmer.janelia.org/) (24) to construct HMM models. Moreover, the hmmsearch program of HMMER 3.0 (24) was used to search all protein sequences in 70 eukaryotic species. The default parameters were chosen for the three tools, and additional filters were used to improve the accuracy (presented in the ‘Discussion’ section). Since one gene can generate multiple variant transcripts, the Ensembl Gene ID was adopted as the unique accession to avoid any redundancy. For one gene, only the protein with the lowest E-value was reserved. All HMM profiles are available at http://uucd.biocuckoo.org/faq.php.
Here, we took 1282 identified E1s, E2s, E3s and DUBs as the benchmark dataset to evaluate the prediction performance and robustness of the HMM identifications. For each family, the annotated sequences were taken as positive data (P), while all other proteins were regarded as negative data (N). The sensitivity (Sn) and specificity (Sp) can be calculated as below:
Both the self-consistency and leave-one-out validations were carried out. The receiver operating characteristic (ROC) curves were illustrated, and AROC (area under ROC) values were calculated for eight families, respectively (Supplementary Figure S1). The results suggested that our predictions are accurate and robust (Supplementary Figure S1). To ensure that all curated proteins can be correctly predicted and classified (Sn = 100%), we selected distinct cut-off values for different families (Supplementary Table S2).
For the families without HMM profiles, we conducted the orthology search for 70 eukaryotes by using the reciprocal best-hit approach (29), which can efficiently identify ortholog pairs if two proteins in two different proteomes reciprocally find each other as the best hit, by the blastall program in the BLAST package (30).
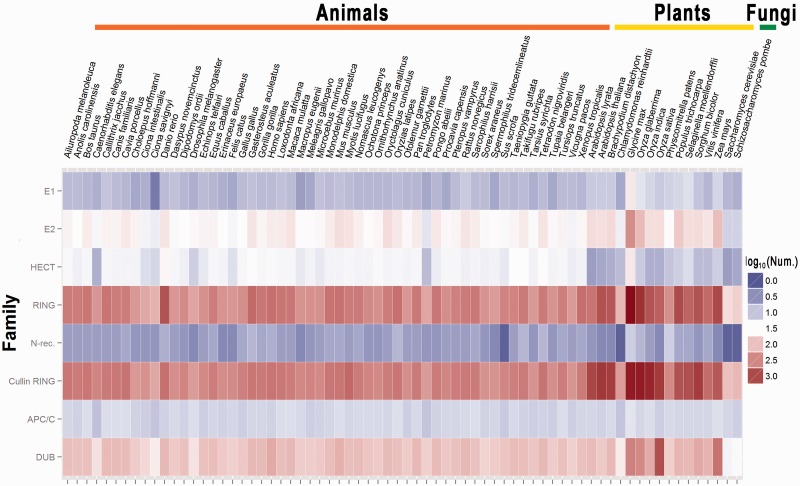
Totally, we computationally characterized 738 E1s, 2937 E2s, 46 631 E3s and 6647 DUBs from 70 eukaryotic species, whereas the heat map of the classifications and results for several major groups or families were illustrated by the ggplot2 program (http://had.co.nz/ggplot2/) in the R package (http://www.r-project.org/) (Figure 1). The detailed classifications and data statistics are available in Supplementary Table S3.
Figure 1.
The heat map of the classifications and protein numbers of E1s, E2s, DUBs and several major groups for E3 ligases.
USAGE
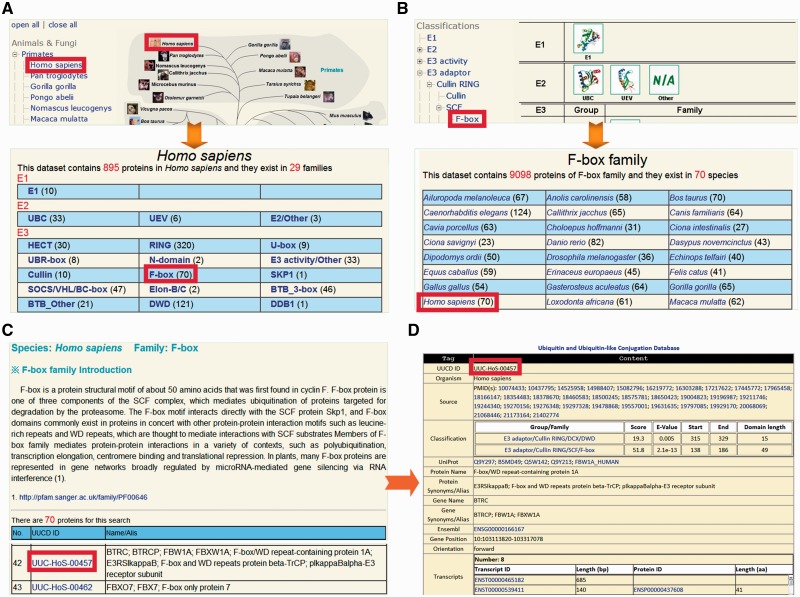
The UUCD database was constructed in an easy-to-use manner. Here, we used human F-box/WD repeat-containing protein 1A (β-TrCP) as an example to describe the usage of UUCD. The Browse page is the major option for users to look through the UUCD conveniently. Two strategies were implemented for browsing the data, including by species and by classifications (Figure 2). In the option of ‘Browse by species’, the right tree represents the phylogenetic relations of eukaryotic species in Ensembl, while the left tree represents the Ensembl taxonomy categories, including primates, rodents, laurasiatheria, afrotheria and so on (25) (Figure 2A). By clicking on the ‘Homo sapiens’ button, the E1, E2, E3 and DUB families in H. sapiens can be shown (Figure 2A). Also, UUCD can be browsed by classifications (Figure 2B). The left tree represents the hierarchical categories, whereas known 3D structures of E1s, E2s, E3s or DUBs were taken from the PDB database (31) and present in right (Figure 2B). Since human β-TrCP belongs to the F-box family, users can click on the ‘F-box’ button to visualize the family information of 70 eukaryotic species (Figure 2B). By either clicking on the ‘F-box’ button in the page of human families (Figure 2A) or the ‘Homo sapiens’ button in the page of the F-box family (Figure 2B), the members in human F-box family can be available, while a brief description is shown for biological functions and regulatory roles of F-box containing proteins (Figure 2C). The UUCD ID (UUC-) was adopted for organizing the database, while the Ensembl Gene ID was used as the secondary accession (Figure 2C). The users can click on the ‘UUC-HoS-00457’ to view the detailed information of human β-TrCP (Figure 2D).
Figure 2.
The browse option of UUCD. We provided two strategies for browsing the database: (A) by species and (B) by classifications. (C) For each family, a brief description and associated members of family were present. (D) The detailed information of human β-TrCP.
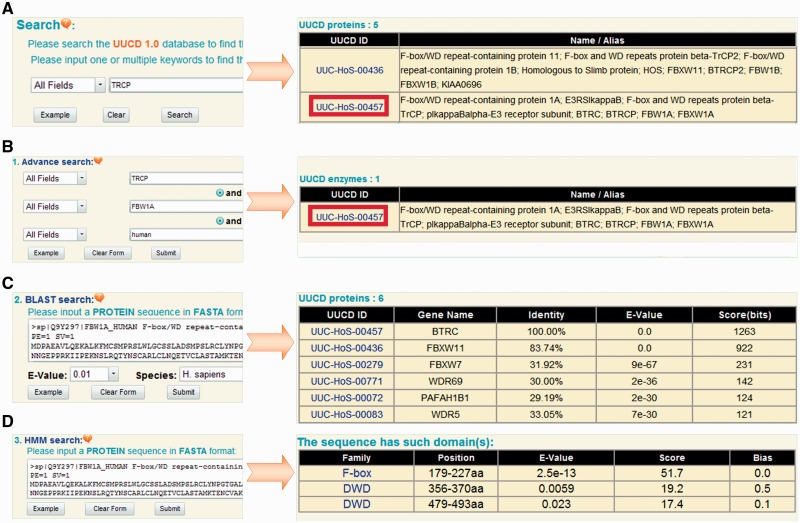
The UUCD can be searched with one or multiple keywords (Figure 3A). For example, if the keyword of ‘TRCP’ is inputted and submitted, the results will be shown in a tabular page, with the features of UUCD ID and protein/gene names/aliases (Figure 3A). Furthermore, three additional advance options were provided, such as (i) advance search, (ii) BLAST search and (iii) HMM search. In advance search option, up to three search terms can be specified and submitted to search the precise information (Figure 3B). The second option, BLAST search, was designed for querying the related information in UUCD by protein sequences. The blastall program of NCBI BLAST packages (30) was integrated in the database. Users can input a protein sequence in FASTA format to search identical or homologous proteins (Figure 3C). (iii) In HMM search option, a protein sequence in FASTA format can be inputted and scanned with 1, 1, 15 and 6 HMM profiles of E1, E2, E3 and DUB families, respectively. If the protein is determined as a ubiquitin-associated enzyme, the classification and detection information will be present (Figure 3D).
Figure 3.
The search and advance options. (A) The database can be queried with one or multiple keywords. (B) Advance search allows users to input up to three terms for the precise search. (C) Blast search option was designed for searching database with one protein sequence in FASTA format. (D) HMM search option will scan the inputted protein sequence with pre-constructed HMM profiles.
DISCUSSION
As one of the most important post-translational modifications of proteins, reversible ubiquitin and ubiquitin-like conjugation have been implicated in almost all aspects of biological processes and functions, and determines the cellular dynamics and plasticity (1,4,5,7–9). Aberrances of ubiquitin and ubiquitin-like conjugation systems are highly implicated in a variety of diseases and cancers (7,10). In this regard, identification and classification of E1s, E2s, E3s and DUBs is fundamental for dissecting the molecular mechanisms and regulatory roles of the conjugation (4,8,9), analyzing the phylogenetic relations of enzymes (8,23), modeling E3-substrate networks (32) and providing potent candidates for drug design (11). Developing a comprehensive database with detailed annotation and classification information has emerged to be an urgent challenge.
Previously, a number of computational efforts were taken for systematically characterizing proteins in the ubiquitin-proteasome system (UPS) (8,13–17). For example, Semple et al. (13) performed a genome-wide study by identifying potential E1s, E2s, E3s and DUBs from four animals and two fungi. From the scientific literature, SCUD (15) and hUbiquitome (17) databases collected experimentally verified enzymes for S. cerevisiae and H. sapiens, respectively. The proteins in the two databases were fully covered by our benchmark dataset (Supplementary Table S1). To the best of our knowledge, currently the most comprehensive database was plantsUPS, which contains predicted E1s, E2s and E3s for up to seven plants, whereas the experimental information was not present (16).
By collecting known enzymes of ubiquitin and ubiquitin-like conjugation systems, we classified them based on distinguishable functional domains (Supplementary Table S3). Although several general databases such as Pfam (27), InterPro (33) and SMART (34) contain most of these domains, we re-constructed HMM profiles with our benchmark dataset to promise the prediction performance, and additional filters were used for accurate classification (Supplementary Figure S1). For example, the HMM profiles of UBC and UEV domains are highly similar and cannot be distinguished by hmmsearch (24). However, the active site cysteine residue locating ∼70–90 aa for ubiquitin coupling does not exist in UEV domains (18) (Supplementary Figure S2). This single rule was adopted for accurately separating UBC and UEV domains (Supplementary Figure S1). Also, the SOCS/VHL/BC-box family is less conserved and cannot be defined with a single domain (35). Thus, proteins containing at least one of SOCS, VHL or BC-box domains were classified into this family. Furthermore, recent analysis revealed a 32 aa ‘3-box’ motif following BTB domain is essential for Cullin binding, although BTB proteins without the motif might interact with Cul3 in alternative mechanisms (36). The proteins predicted with both BTB domain and 3-box motif were classified into the BTB_3-box family (Supplementary Figure S3). In addition, experimental studies suggested that the highly conserved motif EXnHXHX10D is essential for the DUB activity of the JAMM family (37,38). The predicted JAMM proteins without this motif were discarded.
Together with the ortholog search for families without HMM profiles, we systematically identified 738 E1s, 2937 E2s, 46 631 E3s and 6647 DUBs from 70 eukaryotic organisms, with an average number of 813.6 total enzymes per organism (Supplementary Table S3). Although there are 672.5 enzymes encoded in one animal, the average number of plant enzymes is >2-fold (1441.5) higher in animals (Supplementary Table S3). Also, the numbers of animal or plant enzymes in the same group or family can be greatly different (Figure 1). For example, we totally identified 495 E1s in 54 animals with an average number of 9.2 per species, whereas up to 226 E1s were detected in 14 plants with an average number of 16.1 (Supplementary Table S3). However, there were 1803 SOCS/VHL/BC-box proteins identified in animals (33.4 per organism), while only 19 SOCS/VHL/BC-box proteins were detected in plants (Supplementary Table S3). Taken together, our systematic analysis demonstrated the complexity and diversity of enzymes for ubiquitin and ubiquitin-like conjugation in eukaryotes.
For future plans, at least three additional types of ubiquitin-associated proteins will be collected and systematically identified. First, the polyubiquitin ligase (E4) was demonstrated to be responsible for assembling multiubiquitin chains (39). It is still not known how many E4 ligases are encoded in eukaryotic genomes. As the number of known E4s is quite limited, such information was not included in UUCD. Second, accumulative experiments revealed that a large number of ubiquitin-binding domain (UBD)-containing proteins (>150) can non-covalently interact with ubiquitin (40). Thus, UBD proteins can dynamically interact with modified substrates in a ubiquitination-dependent manner and participate in the ubiquitin network (40). Recently, systematic identification of ubiquitinated proteins with modified sites has emerged to be a hot topic (2). Such information will be carefully collected and curated in the near future. Taken together, although more information remains to be integrated, the comprehensive UUCD with E1s, E2s, E3s and DUBs across 70 eukaryotes can serve as a useful resource for further researches. The UUCD database will be continuously updated, when the proteome sequences of more species are available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–3.
FUNDING
Funding for open access charge: The National Basic Research Program (973 Project) [2010CB945401, 2012CB911201, 2012FY112900, 2012CB910101]; National High-tech R&D Program of China (863 Program) [2012AA020401]; National Natural Science Foundation of China [31171263, 81272578, 31071154, 30900835, 30830036, 91019020, 31171271, 31270885].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hershko A. Nobel lecture: the ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle. Angew. Chem. Int. Ed. Engl. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- 2.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl Acad. Sci. USA. 1980;77:1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 5.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 6.Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 7.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat. Rev. Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 11.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc. Natl Acad. Sci. USA. 1981;78:761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semple CA. The comparative proteomics of ubiquitination in mouse. Genome Res. 2003;13:1389–1394. doi: 10.1101/gr.980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee WC, Lee M, Jung JW, Kim KP, Kim D. SCUD: Saccharomyces cerevisiae ubiquitination database. BMC Genomics. 2008;9:440. doi: 10.1186/1471-2164-9-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Z, Zhou X, Li L, Su Z. plantsUPS: a database of plants' Ubiquitin Proteasome System. BMC Genomics. 2009;10:227. doi: 10.1186/1471-2164-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y, Xu N, Lu M, Li T. hUbiquitome: a database of experimentally verified ubiquitination cascades in humans. Database. 2011 doi: 10.1093/database/bar055. Nov 30 (doi: 10.1093/database/bar055; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Wijk SJ, Timmers HT. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24:981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 19.Sriram SM, Kim BY, Kwon YT. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 2011;12:735–747. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- 20.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 21.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 22.Patterson C. A new gun in town: the U box is a ubiquitin ligase domain. Sci. STKE. 2002;2002:pe4. doi: 10.1126/stke.2002.116.pe4. [DOI] [PubMed] [Google Scholar]
- 23.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 25.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 30.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Golovin A, Gore SP, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2012;40:D445–D452. doi: 10.1093/nar/gkr998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y, Lee H, Park JC, Yi GS. E3Net: a system for exploring E3-mediated regulatory networks of cellular functions. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.014076. O111.014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 38.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, III, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 39.Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size' doesn't fit all. Trends Biochem. Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]