Abstract
The Ensembl project (http://www.ensembl.org) provides genome information for sequenced chordate genomes with a particular focus on human, mouse, zebrafish and rat. Our resources include evidenced-based gene sets for all supported species; large-scale whole genome multiple species alignments across vertebrates and clade-specific alignments for eutherian mammals, primates, birds and fish; variation data resources for 17 species and regulation annotations based on ENCODE and other data sets. Ensembl data are accessible through the genome browser at http://www.ensembl.org and through other tools and programmatic interfaces.
INTRODUCTION
Ensembl (http://www.ensembl.org) collects, creates, organizes and distributes data resources in support of research into the genetics and genomics of chordates. We currently support 70 species with a focus on human in additional to agricultural animals and major vertebrate model organisms such as mouse, zebrafish and rat. We support a full range of researchers in genomics from bench biologists interested in looking up specific details about their genes or loci of interest using a graphical web interface to advanced bioinformatics programmers looking to do complex analysis or build new tools that leverage the Ensembl infrastructure. As such, we provide all of the Ensembl source code freely under an Apache-style license and release all of our data without restriction. Ensembl data are distributed from our genome browser at http://www.ensembl.org as well as via BioMart, the Ensembl Application Programming Interface (API), direct MySQL access, Amazon Web Services Public data sets (http://www.ensembl.org/info/data/amazon_aws.html) and via full data download.
Ensembl aims to be a hub of genome information by linking identifiers and information between external biological resources and data within Ensembl or importing essential information from other resources so that it can be found within Ensembl and linked back to the original resource as necessary. For example, we provide up to date external database references to gene names from the HUGO Gene Nomenclature Committee (HGNC) (1), the Universal Protein Resource (UniProt) (2), Orphanet portal for rare diseases and orphan drugs (3), the Online Mendelian Inheritance in Man (OMIM) database (4), the RefSeq collection of Reference Sequences from NCBI (5), the UCSC Genome Browser (6), the Protein Data Bank (PDB) repository for biological macromolecular structures (7) and many other resources.
We participate in or work closely with a number of large-scale international projects including the 1000 Genomes Project (8), ENCODE (9), the International Cancer Genome Consortium (ICGC) (10) and the BLUEPRINT epigenome mapping project (11). Participation in these efforts helps ensure that we produce timely and valuable resources through direct scientific engagement with the communities that we are trying to serve. In addition, we actively develop and provide key pieces of large-scale bioinformatics infrastructure including the eHive workflow management system for genomic analysis (12).
Full incorporation of the data types resulting from the myriad of experimental assays now leveraging next generation sequencing technology remains an important area of development for the project. During the past year, we have made considerable progress in a number of ways including a greater incorporation of RNA-seq data into our gene annotations and ChIP-seq data into our regulatory annotations. In general, we believe that the most useful resources provide integrated summary information that transforms the raw sequencing data into biological knowledge that can provide a foundation for further biological research. Thus, we believe that the display of the called variants from the 1000 Genomes Project or regulatory region annotations supported by specific histone modification or transcription factor (TF) binding sites are more useful as resources for the community than a display of the raw aligned sequence reads. However, Ensembl does support the upload and visualization of read alignment data (e.g. alignment files in BAM format) and provides signal files for our ChIP-seq and alignment files for RNA-seq data within the browser for those users needing direct access to the supporting data. Indeed, Ensembl’s API development this year included increasing support for file-based data access to enable integration of very large BAM and other file-based data sets into the browser.
This report highlights the new data we have released and the new mechanisms of data access that we have deployed during the past year since our previous report (13). We describe how these new features extend the existing capabilities of the project, which will be explained as appropriate.
Supported species
As of release 69 (October 2012), Ensembl supports 70 species including 61 species fully supported on our main site. Of these, we have created full gene annotations for 58 chordates (43 with high-coverage genome sequences and 15 with low-coverage) and have imported annotation data for three non-chordate model organisms (Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster) to facilitate comparative analysis. Five new species were included during the past year with full support: Atlantic cod (Gadus morhua), coelacanth (Latimeria chalumnae), ferret (Mustela putorius furo), Nile tilapia (Oreochromis niloticus) and Chinese softshell turtle (Pelodiscus sinensis). An additional nine species are currently available with limited support on the Ensembl Pre! site (http://pre.ensembl.org) including the following, which were newly added in the past year: budgerigar (Melopsittacus undulates), Chinese hamster CHO cell line (Cricetulus griseus), painted turtle (Chrysemys picta bellii), spotted gar (Lepisosteus oculatus), collared flycatcher (Ficedula albicollis) and squirrel monkey (Saimiri boliviensis boliviensis). Ensembl Pre! sites provide BLAST and genome visualization, but do not provide a complete gene build. For specific genomes, we also provide downloadable data on the preview site.
We update the human gene set for every Ensembl release via a merge of the Ensembl evidence-based automatic annotation and Havana manual annotation (14) to produce an updated GENCODE gene set (9,15). This set also includes all current human Consensus Coding Sequence (CCDS) gene models (16). Manual annotation from Havana is also incorporated into our gene sets on alternate releases for mouse and zebrafish. In addition, pig now includes manual annotation from Havana on selected regions of the genome.
The human genome assembly is updated regularly by the Genome Reference Consortium (GRC) to include alternate sequences in the form of ‘fix’ and ‘novel’ assembly patches (17), and we continue to include these additional alternate sequences and annotate them with genes and other features as appropriate. Ensembl release 69 (October 2012) included GRCh37.p8 (i.e. the eighth patch release of the GRCh37 assembly). The mouse genome annotation, which also incorporates all current mouse CCDS models, was updated for Ensembl release 68 (July 2012) to reflect the new GRCm38 assembly. Other species previously available on our website also saw updates in the past year including new primary assemblies and gene sets for chimpanzee, dog, pig, ground squirrel, bushbaby and Ciona intestinalis. The gene sets for orang-utan, opossum and platypus were also updated using RNA-seq data.
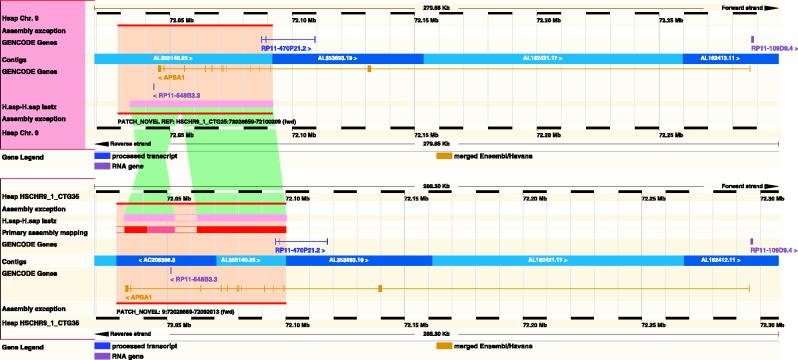
The whole genome multiple and pairwise alignments have been re-run in conjunction with the incorporation of new or updated genomes. In addition to cross-species alignments, we now provide self-alignments for the human genome and also use the Ensembl comparative genomics infrastructure for the comparison of fix and novel patches alongside the reference human genome (Figure 1).
Figure 1.
A region of the GRCh37 human assembly showing the complete APBA1 gene. The top panel displays the GRCh37 reference sequence as originally released, and the bottom panel displays the region after the inclusion of the novel patch HSCHR9_1_CTG35. The region of difference is highlighted and marked by the ‘Assembly exception’ track, whereas the pink regions of LASTZ self-alignment provide more details about what has changed in the patch including the addition of new sequence that was missing in the originally released assembly. The green areas show the mapping between the original and the alternative sequences and demonstrate a corrected inversion at the left hand side of the patch. The patch changes the annotation such that the RNA gene RP11-548B3.3 (in purple) moves from 5′ of the APBA1 gene to within the second intron. As can be seen in the right hand side of the figure, the existence of the patch does not alter the annotation downstream of the change. Figure based on http://e68.ensembl.org/Homo_sapiens/Location/Multi?db=core;r=9:72019177-72298831;r1=HSCHR9_1_CTG35:72019384-72307679;s1=Homo_sapiens–HSCHR9_1_CTG35.
Gene annotation
The year 2012 has seen the inclusion of RNA-seq data provided by several different groups (18–20) as supporting evidence for our gene annotations. Thirteen species currently incorporate RNA-seq data including zebrafish, chimpanzee, Nile tilapia, dog, Chinese softshell turtle, pig, ferret, platyfish, coelacanth, Tasmanian devil, orang-utan, opossum and platypus. For some of these species, the RNA-seq data were added after a standard gene annotation process (21), whereas for other species, the data were added as an integral part of the genebuild process. Some species also include tissue-specific RNA-seq data that enables the exploration of tissue-specific expression. In addition, the Illumina Human BodyMap 2.0 data (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-513) have been re-processed using our enhanced pipeline to produce updated gene models and new BAM files.
RNA-seq data are now routinely used in gene annotation in a number of ways, and we anticipate that RNA-seq data will be used in almost all gene annotation projects for the foreseeable future. Briefly, our current procedure starts with raw-sequencing reads that are aligned to the genome and processed to produce RNA-seq-based gene models, BAM files and intron features that are supported by intron-spanning reads. Intron-supporting evidence helps to quantify intron predictions in RNA-seq transcript sets. The intron features and RNA-seq-based gene models are used alongside cDNA and EST alignments to compare and filter the preliminary set of protein-coding models against a set of highly supported splice sites. In addition, the RNA-seq-based gene models are used to provide alternate isoforms and fill in gaps between models identified by the standard Similarity Genewise component of our annotation system, which aligns protein sequences to the genome, and to add untranslated regions to the protein coding models.
We have also developed an RNA-seq update pipeline that allows an existing Ensembl gene set to be updated through incorporation of new RNA-seq data. The RNA-seq update pipeline takes in the results of the standard Ensembl gene annotation method and also RNA-seq-based models produced by the pipeline previously described (20). The two sets of input models are compared and merged to produce an updated gene set. This new method was used to improve the existing opossum, platypus and orang-tuan gene sets for Ensembl release 69 (October 2012). The method is particularly effective for species that are distantly related to the well-annotated mammals and those with little species-specific sequence data available at the time of initial annotation. Specific improvements from the RNA-seq update pipeline include lengthening truncated genes, merging adjacent gene fragments and splitting artificially merged genes. RNA-seq-based data are also useful for higher primate species that have previously relied largely on human sequence data for annotation, as it allows for the identification of non-human primate-specific gene expression.
Variation resources
We create variation resources for 17 species by importing and merging data from many different sources through our pipeline (22). The current list of variation data is provided at http://www.ensembl.org/info/docs/variation/sources_documentation.html. Most of our SNP and in-del data (rsIDs, locations, allele frequencies and genotypes) come from dbSNP (23). This year, we have updated the Ensembl Variation databases for human, rat, chimpanzee, orang-utan, zebrafish, pig, dog and macaque. We have also remapped the variation data for mouse onto the new GRCm38 assembly before updated GRCm38 mappings were provided by dbSNP and provided the same update for new dog assembly. Available structural variation data have increased considerably, and we have data for human, mouse, horse, zebrafish, cow and macaque largely provided by the DGVa database of copy number and structural variation (24). The human structural variation data are more comprehensive than all other species combined and include >6 million variants of which 5624 are somatic. The variation database infrastructure storing genotypes has also been redeveloped to improve the responsiveness of our displays and to support non-diploid genomes.
The human variation data also include genotypes imported from the 1000 Genomes Project and the NHLBI Exome Sequencing Project (25), ∼79 000 mutation data locations provided by HGMD (26), clinical variants on LRGs (27) and >135 000 somatic mutation positions from COSMIC (28). We have also added mitochondrial variants, information on clinical significance and global minor allele frequencies from dbSNP, as well as phenotype data for >287 000 variants from OMIM (4), the European Genome-phenome Archive (EGA) and the NHGRI GWAS catalog (29). We denote those variants present on three Affymetrix genotyping chips (GeneChip 100 K Array, GeneChip 500 K Array, GenomeWideSNP_6.0) and nine Illumina chips (CytoSNP12v1, Human660W-quad, Human1M-duoV3, CardioMetaboChip, HumanOmni1-Quad, HumanHap650, HumanHap550, HumanOmni2.5 and Human610_Quad), and also indicate those variants curated by UniProt (2).
For all species, we calculate the effect of each variant allele on overlapping Ensembl transcripts and whether the variant falls within an Ensembl regulatory feature, TF binding motif or a high information position within the motif. Our consequence annotation now uses defined Sequence Ontology (SO) terms (30) for all descriptions, which enable querying of ontological relationships in BioMart. More detailed consequence information is also provided for SNPs and in-dels in specific genomic locations such as splice sites. These SO terms have also been adopted by both the UCSC genome browser and ICGC providing a standard to enable easy comparison of variation annotation.
Other resources supporting human variation include calculated linkage disequilibrium values and tag SNPs, in addition to SIFT (31) and PolyPhen (32) predictions for amino acid changes. This year we have switched to using the Ensembl comparative genomics pipeline to provide the ancestral alleles of SNPs and short deletions for human, orang-utan, chimpanzee and macaque (previously this was imported from dbSNP). We have also extensively improved our quality control (QC) procedures, which leverage the eHive software and have been extended to include structural variations.
As a result of our effort to provide the most useful possible summaries of large data sets to our users, we have added new tracks for 1000 Genomes Project common variants and also tracks for each global 1000 Genomes population. Additionally, appropriate phenotype data have been collected into a dedicated section on the Ensembl gene pages. Finally, the documentation section of the website has also been extended and improved for all areas of Ensembl Variation especially for the Variant Effect Predictor (VEP), SO consequences, QC pipeline and API diagrams.
Ensembl web interface
During the past year, development on the Ensembl web interface has continued a combined strategy of small incremental improvements on the website while making substantial progress on a number of major infrastructure-level projects.
On the data display front, we are now able to show alignments of human assembly patches to the reference assembly (Figure 1) and have renamed the ‘Multi-species view’ as ‘Region comparison’ to reflect its wider applicability. We have also added a transcript variation page, similar to the gene variation page but showing only one transcript at a time, which is particularly helpful in the case of large, well-annotated genes that are challenging to display quickly or interpret easily due to their data density. Other additions to the user interface include a new online tool, Region Report, which provides graphical access to the API script of the same name to export sequence, genes and other annotation from one or more regions. We have also re-introduced the ability to save configurations on images: users can turn their choice of tracks on and off and then save this selection in either the browser session or their personal accounts and then quickly return to the same layout at a later time. These configurations can also be grouped into sets (e.g. to combine a set of favourite variation tracks with a set of gene tracks) for even quicker reconfiguration of images.
We have started to refresh the look and feel of the website. For example, our icon set was previously created from various sources and has now been replaced with a single matching set. We have adapted the layout and colour scheme for increased readability, and we are continuing the process of replacing text-heavy pages with simpler, more user-friendly layouts where appropriate.
Finally, major projects nearing completion and scheduled for release by the end of 2012 include a Javascript-based scrollable genome browser called Genoverse that will be incorporated into our location displays for Ensembl release 69 (October 2012) and support for UCSC-style datahubs, which can contain sets of preconfigured tracks or a user-supplied collection of remote resources. Additional work underway includes a top-to-bottom rewrite of our BLAST/BLAT search using the Ensembl eHive job management system supporting a new web frontend, which will be tested on our beta site (http://beta.ensembl.org) before rolling out into a major Ensembl release in 2013.
Regulation
During the past year, we have significantly updated and increased the amount of data available from the Ensembl regulation database. As of Ensembl release 69 (October 2012), there are 532 ChIP-seq and DNase-seq data sets from 13 human and five mouse cell lines. In total, these data sets represent information about the genomic locations of 49 different histone modification types and the binding regions of 113 different TFs. Forty of these TFs have binding matrices available through the JASPAR database (33), and we have incorporated these motif data as positions of high probability TF-binding sites (5% False Discovery Rate) within the binding regions. We have also created a dedicated experimental summary page providing information on individual experimental details and summary metadata, such as references to the raw sequences reads available in the European Nucleotide Archive (34).
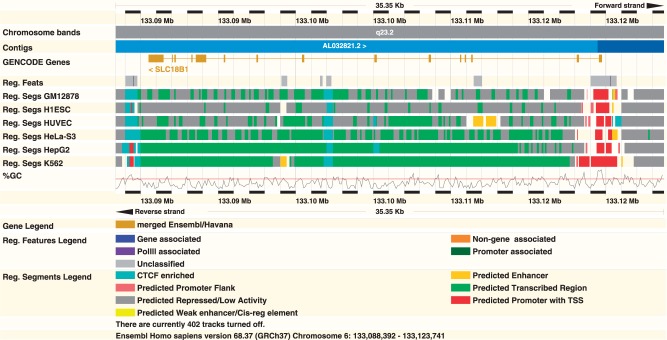
The data underlying the Ensembl Regulatory Build currently include experiments in 13 cell lines. Regulatory Build coverage has increased by 15% in the past year and now annotates 270 Mb of the human genome in 518 020 regulatory features. In Ensembl release 65 (December 2011), we introduced the combined Segway (35) and ChromHMM (36) segmentation analyses developed for ENCODE (9), which classifies the genome into regions based on 12 specific assays to obtain a single-track summary of the functional architecture of the human genome. The segmentation tracks are currently available for six human cell lines: GM12878, K562, H1-hESC, HepG2, HeLa-S3 and HUVEC. The segmentation tracks are displayed with specific views available from the ‘Regulation’ configuration in the Ensembl browser (Figure 2).
Figure 2.
Combined Segway and ChromHMM segmentation analyses within Ensembl in the region around the SLC18B1 gene on human chromosome 6. The combination process results in seven annotated segments: CTCF enriched, Predicted Weak Enhancer/Cis-reg element, Predicted Transcribed Region, Predicted Enhancer, Predicated Promoter Flank, Predicted Repressed/Low Activity or Predicted Promoter with TSS. Six of the seven segment types are shown with variability in predicted enhancer activity between the assayed cell lines. Figure based on http://e68.ensembl.org/Homo_sapiens/Location/View?r=6:133088392-133123741.
The Ensembl Regulation database and web views continue to provide various other data resources including the following: mapping of probe sets for all the common microarray platforms, DNA methylation from various projects including ENCODE, high profile externally curated data sets such as cisRED motifs (37) and an updated VISTA enhancer set (38).
Comparative genomics
New species added in the past year such as coelacanth and lamprey have provided our gene trees with representatives of new taxonomic groups. These species define additional branching points in the phylogenetic trees, enable splitting long branches and provide us with more taxonomic power to better resolve the gene trees. Further information on the evolution of the gene families is now provided by supplementing our phylogenetic analysis with a calculated assessment on the possible expansions and contractions in each family using the CAFE tool (39).
Our data model for gene trees has been modified to handle both protein and ncRNA gene trees. During that process, we also improved our support for protein super-trees, which are used in the resolution of very large protein families. These are split in sub-families, and the super-protein tree represents the relationship between these sub-families. We have developed a better identification and annotation of split genes that usually arise because of assembly errors (40). In our current implementation, the enhanced gene tree pipeline (41) detects gene split events after building the protein multiple alignment, and the resulting nodes of the tree can be annotated as gene split events when they relate to partial proteins that could be concatenated to form a full gene.
Ensembl tools and software
During the past year, we have made significant improvement to the Ensembl VEP (42) and launched a beta implementation of a new Ensembl REST API. The VEP provides comprehensive analysis of SNP, in-del or structural variation data including reports of which gene, transcript, protein or regulatory region overlap the variants of interest and if there is any change in amino acid sequence. It also includes information about SIFT and PolyPHEN predictions in human, protein domains, exon/intron numbers, minor allele frequencies and other information. The VEP works with many different file formats and can in fact convert variant positions between different coordinate systems (Ensembl, RefSeq, LRG and HGVS). We have also written plugins to report on degree of conservation, presence of the variant in an LOVD database in a Locus Specific Database (LSDB) using the Leiden Open Variation Database (LOVD) software (43) and other capabilities. Our VEP plugins are present in the ensembl-variation github repository (https://github.com/ensembl-variation/VEP_plugins), and we encourage users to share their own plugins.
The REST API web service was released as a beta application this year at http://beta.rest.ensembl.org. Although we have a fully supported Perl API to all of the Ensembl data (44), the REST API addresses those users who wish to access Ensembl data in a language-agnostic manner. The web service is built using the Perl web framework Catalyst, Catalyst::Action::REST and our existing Perl API providing a rapid development environment and lowering the cost of creating new endpoints. Output is a combination of bioinformatics and programmatically relevant formats such as FASTA and JSON. We provide access to sequences, assembly mapping, homologues and integration of the VEP with support for genomic features. The REST service, like all Ensembl software, is free to download from our CVS server allowing users to deploy over their local Ensembl databases.
Data access and data mining
Each Ensembl release provides a full rebuild of seven BioMart (45,46) databases. Four of these BioMart databases (Ensembl Gene, Ensembl Variation, Ensembl Regulation and VEGA) are visible on the Ensembl BioMart interface, and the remaining three BioMart databases are hidden from view but are accessed through federation with visible BioMart databases to provide ontology, sequence and genomic feature data. Performing a complete rebuild each release ensures the availability most up to date integrated data from across the Ensembl project. Users can access these data via the MartView (web interface) and MartService (BioMart Perl API, DAS server, SOAP, REST, BioConductor biomaRt package).
Each Ensembl BioMart release includes the addition of any new species, updated assemblies, updates to the germline and somatic variation and structural variation data sets as well as updates to the regulation data. One can now obtain our SIFT and PolyPhen predictions and scores from the Ensembl variation BioMart and from the variation ‘filter’ and ‘attribute’ sections of the Ensembl gene BioMart. It is also possible to select specific mouse strain information from the mouse structural variation data set, and one can filter on the source and study accession of interest in the structural variation data sets available for cow, zebrafish, horse, human, mouse and macaque. A new human somatic structural variation dataset has been added containing data from COSMIC (28). The ability to search multiple chromosomal regions at once has been added to the Ensembl Regulation mart. In addition to this, users can query human regulatory segmentation features using the newly added regulatory segments filter section and attribute page.
User training and support
Ensembl supports new and existing users in a variety of ways from a strong and increasing on-line presence to direct face-to-face training at universities and other institutions worldwide. This year, we held one-day workshops on five continents and launched new virtual initiatives available to all including those further afield or without the means to host a one-day workshop.
We provide extensive free and user-driven tutorials via the Ensembl YouTube (http://www.youtube.com/user/EnsemblHelpdesk) and YouKu (http://i.youku.com/u/id_UMzM1NjkzMTI0) channels and e-learning course (http://www.ebi.ac.uk/training/online/course/ensembl-browsing-chordate-genomes). The Ensembl YouTube channel has >165 subscribers and >91 000 video views, now hosts >20 videos including navigation ‘how-to’ guides. This year, we have added more advanced videos covering subjects such as patches and haplotypes on the human assembly, API installation and how RNA-seq data are used in the genebuild. In 2012, the top 20 countries accessing our on-line training reflect a worldwide audience from the USA, Europe, India, Japan, Australia, Pakistan, Taiwan, Mexico, South Korea and Brazil, and our most popular videos have been viewed hundreds or thousands of times.
We communicate more informally and highlight updates and new features using the Ensembl blog (http://www.ensembl.info/), Facebook page (http://www.facebook.com/Ensembl.org) and Twitter account (http://twitter.com/ensembl). Our Helpdesk (helpdesk@ensembl.org) continues to provide email support for >100 questions monthly, and we are exploring webinars as a vehicle for more interactive long-distance learning and plan to offer more of these events in 2013.
FUNDING
The Wellcome Trust provides majority funding for the Ensembl project [WT062023 and WT079643] with additional funding from the National Human Genome Research Institute [U01HG004695, U54HG004563 and U41HG006104] the BBSRC [BB/I025506/1], and the European Molecular Biology Laboratory. Additional support for specific project components as specified: Funded by the European Commission under SLING, grant agreement number 226073 (Integrating Activity) within Research Infrastructures of the FP7 Capacities Specific Programme; The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no 222664. (“Quantomics”). This Publication reflects only the author's views and the European Community is not liable for any use that may be made of the information contained herein; The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement number 200754 – the GEN2PHEN project; The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/ 2007-2013) under the grant agreement no 223210 CISSTEM; The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement no 282510 – BLUEPRINT. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are consistently grateful to their users and especially to those who take the time to contact us through our mailing lists, blog and other avenues. They acknowledge those researchers, organizations and large-scale projects that have provided data to Ensembl before publication under the understandings of the Fort Lauderdale meeting discussing Community Resource Projects and the Toronto meeting on pre-publication data sharing.
REFERENCES
- 1.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum. Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 4.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(®)) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 5.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Golovin A, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2012;40:D445–D452. doi: 10.1093/nar/gkr998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 12.Severin J, Beal K, Vilella AJ, Fitzgerald S, Schuster M, Gordon L, Ureta-Vidal A, Flicek P, Herrero J. eHive: an artificial intelligence workflow system for genomic analysis. BMC Bioinformatics. 2010;11:240. doi: 10.1186/1471-2105-11-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl.1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harte RA, Farrell CM, Loveland JE, Suner MM, Wilming L, Aken B, Barrell D, Frankish A, Wallin C, et al. Tracking and coordinating an international curation effort for the CCDS Project. Database (Oxford) 2012;2012:bas008. doi: 10.1093/database/bas008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 19.Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, Fu B, Hims M, Ding Z, Ivakhno S, et al. Genome sequencing and analysis of the tasmanian devil and its transmissible cancer. Cell. 2012;148:780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins JE, White S, Searle SM, Stemple DL. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res. 2012;22:2067–2078. doi: 10.1101/gr.137901.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Cunningham F, Rios D, McLaren WM, Smith J, Pritchard B, Spudich GM, Brent S, Kulesha E, et al. Ensembl variation resources. BMC Genomics. 2010;11:293. doi: 10.1186/1471-2164-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foelo ML, Sherry ST. NCBI dbSNP Database: content and searching. In: Weiner MP, Gabriel SB, Stephens JC, editors. Genetic Variation: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 41–61. [Google Scholar]
- 24.Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, Dicuccio M, et al. Public data archives for genomic structural variation. Nat. Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human gene mutation database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 27.Dalgleish R, Flicek P, Cunningham F, Astashyn A, Tully RE, Proctor G, Chen Y, McLaren WM, Larsson P, et al. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The sequence ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 32.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amid C, Birney E, Bower L, Cerdeño-Tárraga A, Cheng Y, Cleland I, Faruque N, Gibson R, Goodgame N, et al. Major submissions tool developments at the European Nucleotide Archive. Nucleic Acids Res. 2012;40:D43–D47. doi: 10.1093/nar/gkr946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson G, Bilenky M, Lin K, He A, Yuen W, Dagpinar M, Varhol R, Teague K, Griffith OL, et al. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 2006;34:D68–D73. doi: 10.1093/nar/gkj075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 40.Dessimoz C, Zoller S, Manousaki T, Qiu H, Meyer A, Kuraku S. Comparative genomics approach to detecting split-coding regions in a low-coverage genome: lessons from the chimaera Callorhinchus milii (Holocephali, Chondrichthyes) Brief Bioinform. 2011;12:474–484. doi: 10.1093/bib/bbr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 44.Stabenau A, McVicker G, Melsopp C, Proctor G, Clamp M, Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]