Abstract
ChEBI (http://www.ebi.ac.uk/chebi) is a database and ontology of chemical entities of biological interest. Over the past few years, ChEBI has continued to grow steadily in content, and has added several new features. In addition to incorporating all user-requested compounds, our annotation efforts have emphasized immunology, natural products and metabolites in many species. All database entries are now ‘is_a’ classified within the ontology, meaning that all of the chemicals are available to semantic reasoning tools that harness the classification hierarchy. We have completely aligned the ontology with the Open Biomedical Ontologies (OBO) Foundry-recommended upper level Basic Formal Ontology. Furthermore, we have aligned our chemical classification with the classification of chemical-involving processes in the Gene Ontology (GO), and as a result of this effort, the majority of chemical-involving processes in GO are now defined in terms of the ChEBI entities that participate in them. This effort necessitated incorporating many additional biologically relevant compounds. We have incorporated additional data types including reference citations, and the species and component for metabolites. Finally, our website and web services have had several enhancements, most notably the provision of a dynamic new interactive graph-based ontology visualization.
INTRODUCTION
Chemical Entities of Biological Interest (ChEBI) is a database and ontology for low-molecular-weight chemical entities that are relevant for understanding and intervening in biological functioning (1,2). The core information ChEBI provides is a stable and unique ChEBI identifier along with a name, chemical structure and ontological classification. A wide variety of additional information is provided to enhance this core offering, including International Union of Pure and Applied Chemistry (IUPAC) recommended name and other synonyms, chemical data such as mass and formula, cross-references to other databases and references such as Wikipedia, applications and biological roles, and citations relevant to that chemical entity. Each entry in the database is manually annotated by expert annotators before being incorporated into a monthly release. All data are checked against the primary literature and other publicly available data resources. Chemical entities are classified in the ontology taxonomically, in terms of their main structural features. They are also assigned one or more roles based on their activities or uses in biological contexts. Chemical–role relationships can be variably sourced, including from the primary literature, textbooks, information contained in other publicly available databases, and curatorial knowledge.
External databases, including the Rhea database of biochemical reactions (3), the Reactome database of biological pathways and processes (4) and the BioModels database of systems biology mathematical models (5), use the stable and unique primary ChEBI identifiers to refer unambiguously to the chemicals as they appear in their biological context. ChEBI is also used as the chemical reference to provide logical definitions for chemical-involving biological entities by other ontologies such as the Gene Ontology (6,7).
Since our earlier reports on ChEBI (1,2), the database has steadily grown, and we have introduced several new features and enhancements. We report here exclusively on these new aspects of ChEBI, and refer the interested reader to the previous reports for the description of features that have not been recently introduced.
DATABASE CONTENT EVOLUTION
Over the past few years, ChEBI has steadily grown in annotated content, with the November 2012 release containing nearly 30 000 fully annotated entities. For a more detailed overview, the current state of the data content in ChEBI is broken down by entity type and level of curation in Table 1.
Table 1.
ChEBI data content
| Curation level | Chemical classes | Compounds | Roles | Subatomic particles | All ChEBI | |
|---|---|---|---|---|---|---|
| Number of entries | 1 | 2352 | 1201 | 113 | 1 | 3667 |
| 2 | 40 | 4321 | 0 | 0 | 4361 | |
| 3 | 8128 | 20 961 | 652 | 41 | 29 782 | |
| ALL | 10 520 | 26 483 | 765 | 42 | 37 810 | |
| Number with definitions | 1 | 116 | 262 | 40 | 1 | 419 |
| 2 | 14 | 4316 | 0 | 0 | 4330 | |
| 3 | 4215 | 11 763 | 601 | 38 | 16 617 | |
| ALL | 4345 | 16 341 | 641 | 39 | 21 366 | |
| Number of relationships | 1 | 2352 | 1201 | 113 | 1 | 11 185 |
| 2 | 40 | 4321 | 0 | 0 | 8936 | |
| 3 | 8128 | 20 961 | 652 | 41 | 149 680 | |
| ALL | 10 520 | 26 483 | 765 | 42 | 169 801 | |
| Number of ‘is_a’ relationships | 1 | 6802 | 2749 | 202 | 1 | 9754 |
| 2 | 59 | 4462 | 0 | 0 | 4521 | |
| 3 | 55 739 | 33 645 | 1496 | 98 | 90 978 | |
| ALL | 62 600 | 40 856 | 1698 | 99 | 105 253 | |
| Number of ‘has_role’ relationships | 1 | 80 | 257 | 292 | 0 | 629 |
| 2 | 2 | 4377 | 0 | 0 | 4379 | |
| 3 | 486 | 7690 | 12 596 | 0 | 20 772 | |
| ALL | 568 | 12 324 | 12 888 | 0 | 25 780 | |
| Number of ‘has_part’ relationships | 1 | 59 | 11 | 0 | 0 | 70 |
| 2 | 4 | 0 | 0 | 0 | 4 | |
| 3 | 1044 | 2269 | 8 | 15 | 3336 | |
| ALL | 1107 | 2280 | 8 | 15 | 3410 | |
| Number of structural relationships | 1 | 412 | 320 | 0 | 0 | 732 |
| 2 | 26 | 6 | 0 | 0 | 32 | |
| 3 | 4064 | 30 530 | 0 | 0 | 34 594 | |
| ALL | 4502 | 30 856 | 0 | 0 | 35 358 | |
| Number of citations | 1 | 32 | 387 | 0 | 0 | 419 |
| 2 | 11 | 4510 | 0 | 0 | 4521 | |
| 3 | 1734 | 17 604 | 38 | 1 | 19 377 | |
| ALL | 1777 | 22 501 | 38 | 1 | 24 317 | |
| Number of cross-references | 1 | 1672 | 1440 | 6 | 0 | 3118 |
| 2 | 3 | 3151 | 0 | 0 | 3154 | |
| 3 | 2458 | 52 978 | 104 | 11 | 55 551 | |
| ALL | 4133 | 57 569 | 110 | 11 | 61 823 |
This table shows a breakdown of the ChEBI data content in November 2012, by type of entity and by level of curation. The types of entity are chemical classes (grouping classes in the structure-based classification), compounds (fully specified with chemical structure) and roles. The levels of curation are as follows: 1–barely curated; 2–lightly curated; and 3–fully curated.
We have seen a steady increase in the number of submissions created directly in the database by our users using our submission tool introduced in 2009 (2). More than 2400 of the current total of fully annotated ChEBI entities have been directly submitted using this tool, and the rate of new submissions has seen continuous growth. The ChEBI database content is reported on with every release within the Statistics page (http://www.ebi.ac.uk/chebi/statisticsForward.do). Future work will incorporate content updates (for example, the addition of cross-references to existing entries) into this view.
A large-scale ongoing curation effort in collaboration with the La Jolla Institute for Allergy and Immunology (LIAI, http://www.liai.org/) is focused on annotating compounds relevant for immunology, such as those that act as antigens and immunogens. ChEBI has so far annotated >3800 such compounds. The compounds that are relevant for immunology are provided with roles by the LIAI annotators, and those roles are linked to the relevant compounds in ChEBI using the ‘has_role’ relationship.
Natural products are of substantial interest in drug discovery and metabolism research, as they represent molecules that in many cases have been shaped by natural selection to be bioactive in highly specific ways. ChEBI and partner database MetaboLights (http://www.ebi.ac.uk/metabolights/) are aiming together to become a comprehensive public open resource for describing natural product chemistry. Working towards this objective, ChEBI has recently added >4000 natural products, of which >150 have thus far been fully curated. Together with the pre-existing metabolites in ChEBI, this brings the total collection of metabolites (including both primary and secondary metabolites) close to 5000 (November 2012).
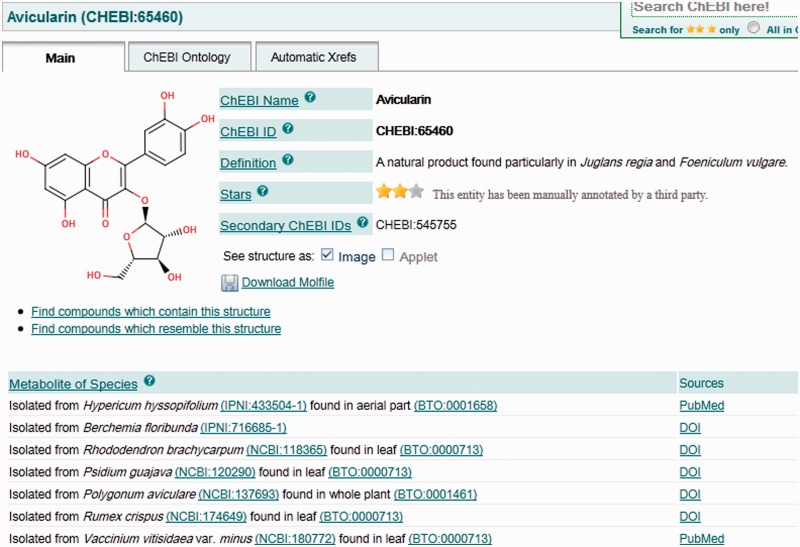
These natural products together with their metabolite species information have been extracted from the primary literature, specifically publications that report identification of the particular metabolite in a given species. The relevant citations are linked to the compound entry as a whole and to the species annotation in particular. To support the representation of this information, several new features and data items have been added. These include fields for storing and searching the source information, such as species, strain and component (tissue type), as well as links to the appropriate taxonomies and ontologies, together with citations for this information in the primary literature. This is illustrated in Figure 1. The linking of specific citations to specific data points in this way is a novel feature in ChEBI but is an approach that we expect to make extensive use of in the future.
Figure 1.
CHEBI:65460 is an example of a natural product recently introduced into ChEBI. It has been isolated from several different species, information about which is included in the new ‘Metabolite of Species’ section of the ChEBI web interface. The information about species and component is linked directly to the reference which acts as a source for that information.
In addition to the creation of new entries in ChEBI, significant effort has been put into updating and enhancing existing entries. This has been done both on an individual record basis (e.g., as a result of a user request) and in a more systematic way through programmatic updating of batches of selected compounds. For example, information on the biological roles of drugs obtained from DrugBank (8) has been harnessed and added where appropriate to the records for drugs already included in ChEBI. Aside from citations mentioning species, ChEBI entries have been associated with general citations. These are manually annotated to support and enhance any aspect of the ChEBI data provision. As of the November 2012 release, 10 512 ChEBI entries are associated with at least one citation, and 1023 with at least five citations. 21 366 ChEBI entries have annotated textual definitions. Text definitions are manually annotated by the ChEBI curation team, focusing on the specific features that differentiate the compound from the immediately asserted parents in the taxonomic hierarchy. These definitions are intended to help non-chemists make optimal use of the ChEBI ontology even when they may not wish to interpret the details of the chemical structures.
We have added database links to several new resources including MetaCyc (9), ChemSpider (10) and Wikipedia. In addition, we now annotate Reaxys registry numbers, where appropriate, in addition to the older Beilstein registry numbers. 23 165 ChEBI entries are associated with at least one manually annotated database link, whereas 3774 are associated with at least five database links. As a convenience feature for our users, the Wikipedia cross-references are also now used to introduce a text box on our entry page that displays the information contained in the first paragraph of Wikipedia for that entity.
WEB INTERFACE AND VISUALIZATION
User experience studies and studies with the real users doing searches and other interactions with the ChEBI interface were conducted, and as a result several changes and simplifications to the ChEBI interface have been introduced, many of which were quite straightforward from a technical perspective but which we believe have led to a better user experience overall.
The studies showed that many users were not clear about the difference between a substructure search and a similarity search. Simply changing the labels for these searches to ‘Find compounds which contain this structure’ and ‘Find compounds which resemble this structure’, respectively, significantly enhanced users’ understanding of the searches being performed. Many users complained that the size of the box used to display search answers meant that many structures were difficult or impossible to distinguish from one another, but still wanted to see as many search answers as possible on the screen. By hovering the cursor over a structure, an enlarged image of the structure now appears on the screen, disappearing again when the cursor is moved away from the structure. A similar feature has also been introduced for the structure on the display page for all ChEBI entries.
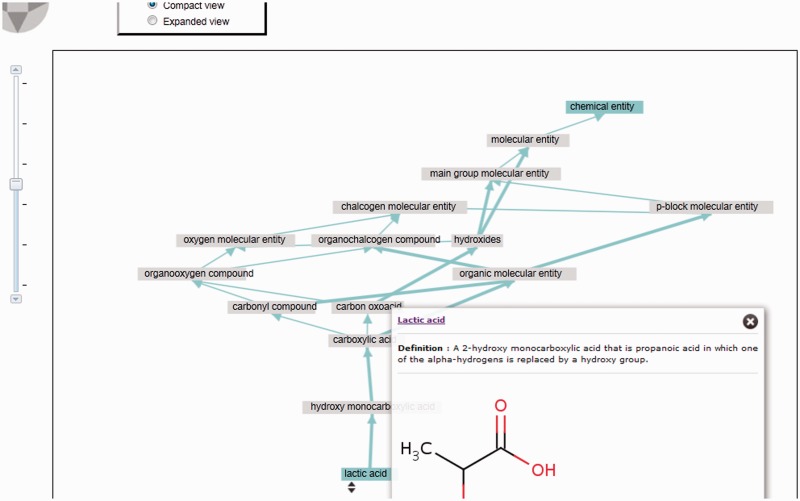
Although experienced users liked the ‘tree view’ of the ChEBI ontology, many others found it to be confusing and intimidating. Following a major redesign of the ontology display, most of the ontology information has been removed from the main display page for a compound, and a new ontology tab has been introduced. Here, the ‘is_a’ and ‘has_part’ relationships are shown in a fully interactive graphical display, illustrated in Figure 2.
Figure 2.
The newly introduced interactive graph-based ontology visualization shows paths from any ontology entity up to the root of the ontology. It supports two views: a compact view (the default, illustrated here) and an expanded view. In the compact view, paths traversing intervening entities for which no additional branching is introduced are suppressed (shown by a thicker line). These can be individually expanded by clicking on a thick line. The figure can be zoomed and any node can be dragged. Tooltips appear when a node is clicked on, giving the definition and other relevant information for that entity.
The interactive graph-based ontology visualization shows the diverse ‘is_a’ paths from the selected ontology entity up to the root of the ontology, which will be one of ‘chemical entity’, ‘role’ and ‘subatomic particle’, depending on the nature of the entity. It also displays relevant ‘has_part’ relationships (as black coloured edges), although without including all the ancestors of the target of the parthood relation, in order to ensure that the information displayed does not become overwhelmed by the proliferation of parts common to most or all chemical entities. The component is based on the ‘force-directed’ graph layout component from the JavaScript InfoVis toolkit (http://thejit.org/), with modifications to customize the display to meet the ChEBI-specific requirements. The figure can be zoomed and any node can be dragged to a preferred position. Tooltips appear when a node is clicked on, giving the definition and other relevant information for that entity.
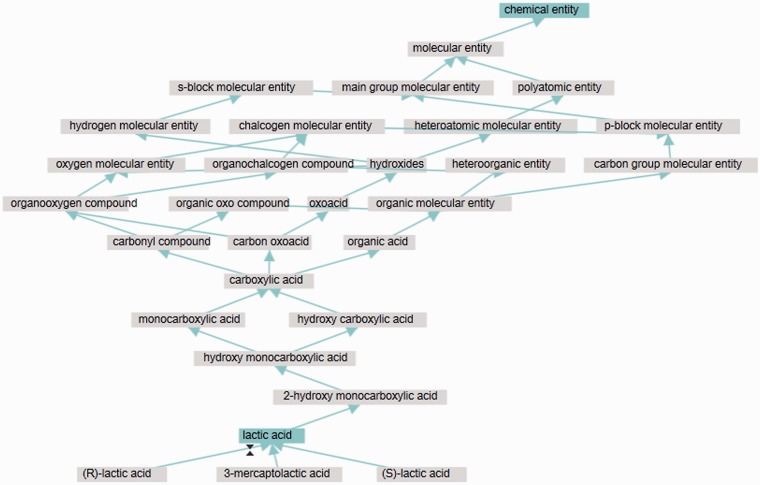
The visualization supports two views: a compact view (the default), illustrated in Figure 2, and an expanded view, illustrated in Figure 3. In the compact view, paths traversing intervening entities for which no additional branching is introduced are suppressed (shown by a thicker line). These can be individually expanded by clicking on a selected thick line to display hidden nodes. In addition, ‘is_a’ children of the selected entity are not displayed in the compact view. These features were introduced to reduce the complexity of the display, while still preserving the most relevant information. The resulting graph display can be compared with the display used in, for example, the OLSVis visualization (11). In OLSVis, all relationship types are illustrated, and all ancestors are included. For many ChEBI entities, however, the result is simply too detailed and overwhelming to be useful for many casual browsers.
Figure 3.
In the expanded view of the interactive ontology visualization, all ancestor nodes and the immediate children of the selected entity are displayed. The other features of the display all remain unchanged.
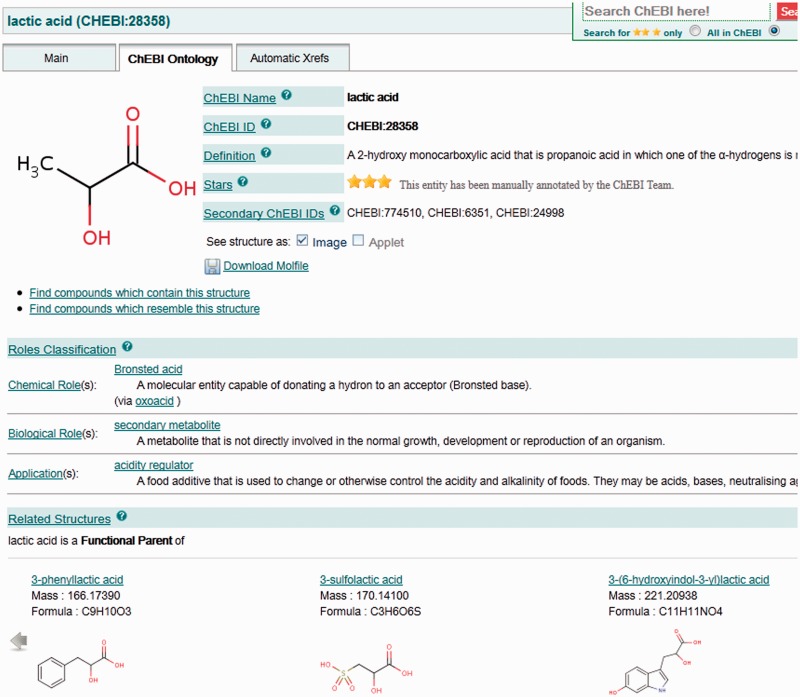
ChEBI has taken the approach of listing the roles and the structural relationships (e.g. ‘is_enantiomer_of’) separately to the graph-based browser. These are displayed along with the structures of the related entities in the ‘Related Structures’ section of the dedicated ontology view, as illustrated in Figure 4. Biological and chemical role terms applying to the entity are also listed separately, along with the definition of each role. The ‘tree view’ remains available from the ontology page as an alternative display.
Figure 4.
Inherited role classifications and chemical structural relationships are illustrated separately on the newly introduced ‘ChEBI Ontology’ tab, above the graph and tree view components. Where the entity being browsed is a role, the list of compounds having that role directly asserted or inherited is displayed in this section, showing the structures of the members and allowing navigation through large results by scrolling.
ONTOLOGY DEVELOPMENT
Accompanying the focus on natural products as part of our curation effort, ChEBI evaluated the classification of natural products within the ontology. An initial challenge was the core definition of natural products, i.e. what specifically constitutes a ‘natural product’. The term is actively used by diverse communities, but its intended meaning is seldom rendered explicit. Some candidate definitions among the many possibilities, in sequence from more inclusive to more exclusive, are as follows:
All chemicals that can be isolated from a living organism;
All metabolites (primary and secondary);
Secondary metabolites only;
Secondary metabolites in plants only.
Of these options, ChEBI has elected to favour the third: thus ‘secondary metabolite’ is now explicitly asserted to have the synonym ‘natural product’ in ChEBI. Furthermore, ChEBI previously included classes related to natural products in two different places within the ontology. Firstly, common natural product families were explicitly classified in the chemical entity ontology, while secondly, ‘metabolite’ was specified in the role ontology. An example of a natural product class that was previously classified in the chemical entity ontology is cinchonine, as ‘is_a’ heterocyclic natural product ‘is_a’ natural product. For clarity, inference capability and ease of searching, this double classification practice has been discontinued.
To comply with our goal of increasing interoperability with other ontologies in the biomedical domain, ChEBI has provided a mapping to the upper level ontology Basic Formal Ontology (BFO) (12), version 2.0. The ChEBI-BFO mapping is provided as a bridge OWL [Web Ontology Language, version 2 (13)] file, downloadable alongside the ChEBI ontology OWL export available at ftp://ftp.ebi.ac.uk/pub/databases/chebi/ontology/. Another extension of ChEBI, introduced as a downloadable OWL file in the same directory, is the assertion of disjoint relationships between high-level entities such as between ‘molecular entity’ and ‘group’. The assertion of disjoints allows the use of an OWL reasoner to automatically detect errors in the asserted ontology. For example, using the reasoner we were able to detect and resolve a number of legacy errors. A major annotation push has ensured that all database entries are now ‘is_a’ classified within the ontology, meaning that all of the chemicals are available to semantic reasoning tools that harness the classification hierarchy.
A further change within the ontology has been the partitioning of all entities originally classified as carbohydrates into one of two disjoint classes, ‘carbohydrate’ and ‘carbohydrate derivative’. As a result, classes such as ‘nucleotide’ in ChEBI, which describe entities that contain a carbohydrate part-structure, can now be classified in a way that is acceptable to both chemists and biologists. This was one of the changes introduced into ChEBI as part of its alignment to the Gene Ontology, which has been used to partially automate the classification of chemical-involving processes in GO based on the ChEBI hierarchy (7).
To deal adequately with user-requested mixtures and polymers within the ontology, ChEBI has introduced a high-level ‘chemical substance’ ontology term in the chemical entity hierarchy, which is in turn further differentiated between pure and mixed substances. A pure substance is a macroscopic homogeneous collection of molecular entities, whereas a mixture includes at least two different types of molecular entity. This allows us to adequately model racemic mixtures, which are crucial in the representation of drugs.
DISCUSSION AND CONCLUSION
High-quality reference chemical data are core to integration, annotation, discovery and analysis activities throughout many sub-disciplines of biological research (14,15). In silico research in domains such as predictive toxicology, metabolic modelling and the search for biomarkers for diseases requires high-quality chemical reference data about the molecular entities that appear in and intervene with the functioning of living organisms. For example, in systems biology modelling, the use of unambiguous, publicly available and perennial identifiers for model components is becoming increasingly recognized as being essential for sharing and reusing models within communities (16). Furthermore, recognized semantic terms are required if such models are to be used in computational pipelines, allowing for the automation of the systems biology life cycle, from model generation, through to integration with experimental omics data, and simulation (17). Ontologies such as ChEBI are essential to making the semantics of such reference terminology computable and useful in algorithms and applications. Other applications that have made use of the semantic information encoded in ChEBI include semantic similarity for improved classification and text mining (18), enrichment analysis using ChEBI biological roles and applications (19) and the establishment of links between drugs and pathways (20). The inclusion of additional data for natural products in ChEBI is of benefit to many application areas including academic drug discovery, for which the provision of such data to the public domain enhances the capability of open data-backed metrics such as natural-product-likeness scores (21).
As mentioned above, ChEBI has already been adopted as the primary chemical annotation and identification standard in several biological databases and ontologies, including BioModels (5), Reactome (4) and the Gene Ontology (7). ChEBI is also listed as a secondary identifier for chemical information in many additional databases such as KEGG (22), DrugBank (8) and HMDB (23).
All ChEBI data are available in PubChem (24), and in a recent release, PubChem have incorporated classification information from the ChEBI ontology into their newly introduced ‘Classification’ sub-section of compound entry pages. To facilitate access to this information, our PubChem cross-references, along with other automatically aggregated cross-references to resources such as UniProt (25), IntAct (26) and SABIO-RK (27), are available on the ‘Automatic Xrefs’ tab of the ChEBI web display and are updated with every monthly release. However, other cross-references, including MetaCyc, HMDB and KEGG, are manually annotated and checked in ChEBI as part of the curation process, and therefore growth in coverage in these database links is slower. We are developing methods to semi-automatically supplement the manual cross-reference annotation. These methods have already been used to supplement the DrugBank links, and will soon be applied to HMDB, KEGG and MetaCyc.
With the enhancements we have here reported on, ChEBI continues in its evolution as a user-driven resource aiming to meet the requirements for chemical knowledge arising from an increasingly diverse group of biological communities.
FUNDING
European Commission [FELICS 021902, SLING 226073]; Biotechnology and Biological Sciences Research Council [BB/G022747/1]. Funding for the open access charge: European Commission [SLING 226073].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are extremely grateful for the continuing contributions from many members of the scientific community to the success of ChEBI. In particular, we would like to thank Randi Vita, Bjoern Peters, Kristian Axelsen, Alan Bridge, Anne Morgat, Colin Batchelor, Neil Swainston, Pedro Mendes, Jane Lomax, Rebecca Foulger, Paola Roncaglia, Adam Bernard, Pablo Moreno, John May, Kenneth Haug, Nicolas le Novère, Barry Smith and all active ChEBI submitters. We also gratefully acknowledge the software contributions of ChemAxon [https://www.chemaxon.com/products/marvin/].
REFERENCES
- 1.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcántara R, Axelsen KB, Morgat A, Belda E, Coudert E, Bridge A, Cao H, de Matos P, Ennis M, Turner S, et al. Rhea—a manually curated resource of biochemical reactions. Nucleic Acids Res. 2011;40:D754–D760. doi: 10.1093/nar/gkr1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, Chelliah V, Li L, He E, Henry A, Stefan M, et al. BioModels database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst. Biol. 2010;4:92. doi: 10.1186/1752-0509-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The GO Consortium. The gene ontology: enhancements for 2011. Nucleic Acids Res. 2011;40:D559–D664. doi: 10.1093/nar/gkr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wishart D, Knox C, Guo A, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams A. ChemSpider and its expanding web: Building a structure-centric community for chemists. Chem. Int. 2008;30:1. [Google Scholar]
- 11.Vercruysse S, Venkatesan A, Kuiper M. OLSVis: an animated, interactive visual browser for bio-ontologies. BMC Bioinformatics. 2012;13:116. doi: 10.1186/1471-2105-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenon P, Smith B. SNAP and SPAN: Towards dynamic spatial ontology. Spatial Cognit. Comput. 2004;4:69–104. [Google Scholar]
- 13.Grau BC, Horrocks I, Motik B, Parsia B, Patel-Schneider P, Sattler U. OWL 2: the next step for OWL. Web Semantics. 2008;6:309–322. [Google Scholar]
- 14.Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- 15.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Swainston N, Smallbone K, Mendes P, Kell DB, Paton NW. The SuBliMinaL Toolbox: automating steps in the reconstruction of metabolic networks. J. Integr. Bioinform. 2011;8:186. doi: 10.2390/biecoll-jib-2011-186. [DOI] [PubMed] [Google Scholar]
- 17.Courtot M, Juty N, Knüpfer C, Waltemath D, Zhukova A, Dräger A, Dumontier M, Finney A, Golebiewski M, Hastings J, et al. Controlled vocabularies and semantics in systems biology. Mol. Syst. Biol. 2011;7:543. doi: 10.1038/msb.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira JaD, Couto FM. Semantic similarity for automatic classification of chemical compounds. PLoS Comput. Biol. 2010;6:e1000937. doi: 10.1371/journal.pcbi.1000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chagoyen M, Pazos F. MBRole: enrichment analysis of metabolomic data. Bioinformatics. 2011;27:730–731. doi: 10.1093/bioinformatics/btr001. [DOI] [PubMed] [Google Scholar]
- 20.Hoehndorf R, Dumontier M, Gkoutos GV. Identifying aberrant pathways through integrated analysis of knowledge in pharmacogenomics. Bioinformatics. 2012;28:2169–2175. doi: 10.1093/bioinformatics/bts350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayaseelan KV, Moreno P, Truszkowski A, Ertl P, Steinbeck C. Natural product-likeness score revisited: an open-source, open-data implementation. BMC Bioinformatics. 2012;13:106. doi: 10.1186/1471-2105-13-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita K, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, Knox C, Guo ACC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. January 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolton E, Wang Y, Thiessen PA, Bryant SH. Washington, DC: American Chemical Society; 2008. PubChem: integrated platform of small molecules and biological activities. [Google Scholar]
- 25.Magrane M, Uniprot Consortium. UniProt knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35: D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittig U, Golebiewski M, Kania R, Krebs O, Mir S, Weidemann A, Anstein S, Saric J, Rojas I. In: Proceedings of the 3rd International workshop on Data Integration in the Life Sciences 2006 (DILS’06) Hinxton, UK: 2006. SABIO-RK: integration and curation of reaction kinetics data. Lect. Notes Bioinformatics, 4075, 94–103. [Google Scholar]