Abstract
The Eukaryotic Promoter Database (EPD), available online at http://epd.vital-it.ch, is a collection of experimentally defined eukaryotic POL II promoters which has been maintained for more than 25 years. A promoter is represented by a single position in the genome, typically the major transcription start site (TSS). EPD primarily serves biologists interested in analysing the motif content, chromatin structure or DNA methylation status of co-regulated promoter subsets. Initially, promoter evidence came from TSS mapping experiments targeted at single genes and published in journal articles. Today, the TSS positions provided by EPD are inferred from next-generation sequencing data distributed in electronic form. Traditionally, EPD has been a high-quality database with low coverage. The focus of recent efforts has been to reach complete gene coverage for important model organisms. To this end, we introduced a new section called EPDnew, which is automatically assembled from multiple, carefully selected input datasets. As another novelty, we started to use chromatin signatures in addition to mRNA 5′tags to locate promoters of weekly expressed genes. Regarding user interfaces, we introduced a new promoter viewer which enables users to explore promoter-defining experimental evidence in a UCSC genome browser window.
INTRODUCTION
The Eukaryotic Promoter Database (EPD) is an old database that has been maintained for more than 25 years. Initially it was based on two principles: (i) promoters were conceptually defined as transcription start sites (TSSs); and (ii) information was gathered by an independent and critical analysis of results published in journal articles. EPD was successfully maintained according to these guidelines for 15 years at least. A comprehensive description of the contents and format of the original EPD database can be found in (1).
Starting about 10 years ago, there was a gradual change in the way TSSs were mapped by experimental researchers. New technologies appeared, including high-throughput sequencing of oligo-capped cDNAs (2) and CAGE (3), which allow for comprehensive characterization of mRNA 5′-ends of a whole transcriptome at once. The DDBJ and EMBL nucleotide sequence libraries introduced a new division called MGA (Mass sequences for Genome Annotation) specifically for this type of data (4). We reacted to this trend by introducing semi-automatic procedures for inferring promoter positions (5) using a new algorithm for TSS clustering (6).
Today, so-called epigenetic profiling assays with an even higher throughput and based on next-generation sequencing (NGS) technologies are confronting us with new challenges and opportunities. For instance, the ChIP-Seq technique (7) targeted at modified histones and components of the transcription machinery reveals the structure and physiological state of chromatin with unprecedented detail, near-basepair resolution and on a genome-wide scale. Likewise, BS-Seq allows for a comprehensive characterization of the DNA methylome (8). More details on such methods can be found in (9).
The wide-spread application of epigenetic profiling techniques led to major new insights about transcriptional regulation and promoter structure (10,11) which prompted us to revise EPD’s underlying promoter definition. In this respect, the increasingly recognized wide-spread occurrence of promoters with dispersed initiation site patterns (12) poses no problem as the schema of EPD always distinguished between three classes of transcription initiation patterns: single sites, clustered multiple sites and transcription initiation regions (1). However, recent findings about promoter histone modifications, including our work on nucleosome architecture (13), suggest that histone modification data should become part of an experimental promoter definition (in addition to the so far exclusively used TSS mapping data). In fact, it has been argued that transcriptional initiation is a fuzzy and somewhat tissue-specific process whereas the formation of a pre-initiation complex and its induced chromatin organization is a more precisely localized and tissue-invariant event.
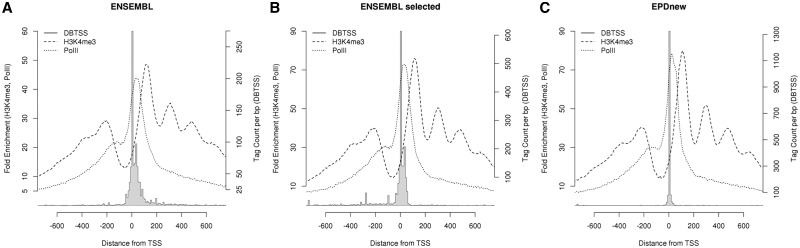
A consensus chromatin signature of a human promoter is shown in Figure 1. The peaks in the H3K4me3 profiles correspond to nucleosome center positions. Hallmarks of a promoter are: (i) a nucleosome-free region around and upstream of the TSS; (ii) a Pol-II peak slightly downstream of the TSS and possibly corresponding to a paused RNA polymerase; and (iii) several positioned, H3K4me3-marked nucleosomes in the promoter downstream region. In response to these findings, we have started to use ChIP-Seq data for defining promoter positions in the latest version of EPD. In essence this amounts to a replacement of the old, transcription initiation site-centric promoter definition by a new composite definition based on a multi-faceted promoter concept.
Figure 1.
Positional distribution of TSSs and selected chromatin marks in an ENSEMBL-derived human promoter set (A), in the subset of ENSEMBL promoters that was selected for inclusion in EPD (B), and in promoters from EPDnew where TSS positions where re-assigned with the aid of new CAGE and oligo-capping data (C). TSS tags corresponding to the 5′-ends of oligo-capped cDNAs were taken fromDBTSS version 7 (14). ChIP-Seq data for H3K4me3 and Pol-II were taken from (7).
In order to exploit the full potential of NGS data, we felt that a complete re-design of EPD was necessary. We therefore added in 2011 a new section called EPDnew, which consists of organism-specific TSS collections automatically assembled from carefully selected MGA data. EPDnew is expected to gradually replace the old corpus of manually curated promoter entries over time. The following section of this article mainly presents the design principles, data acquisition methods, quality control procedures and user interfaces of EPDnew.
RECENT DEVELOPMENTS
Design principles of EPDnew
In essence, the scope of EPD remains unchanged. The specific objectives of EPDnew are: (i) to provide comprehensive and high-quality promoter collections for a number of eukaryotic model organisms; and (ii) to provide integrated views of promoter-defining evidence and other promoter-relevant information by means of custom track files that can be viewed in a UCSC genome browser (15) window. As in the past, we provide for each promoter a single, carefully chosen reference TSS position in order to support regulatory sequence analysis platforms such as RSAT (16) or GREAT (17) which rely on single-base TSS annotation for promoter sequence extraction. In addition, we continue to classify each promoter as ‘single’, ‘multiple’ or ‘region’ according to the spread of the initiation sites pattern. On the other hand, providing regulatory information on individual promoters is not a focus at the moment.
During an initial period, we will focus on known protein coding genes only, for which we try to reach complete coverage as soon as possible. For this reason, we are currently not exploiting new transcriptome data resources for de novo discovery of new promoters. The main difference to the old part of EPD is that the new promoter collections are generated from scratch by an automatic procedure taking primary experimental data and ENSEMBL gene annotation as input. Practically this means that individual entries are no longer propagated from one release to the next. This issue may require some explanations. It is important to recognize that the automatic assembly pipeline generating EPDnew may assign any number of promoters to a given gene. Therefore, if the number of promoters per gene changes, it will not be possible to match promoters in the new version to promoters in the old version. On the positive side, the de novo generation approach ensures that gene names in EPD are always in sync with the current version of the gene nomenclature resource.
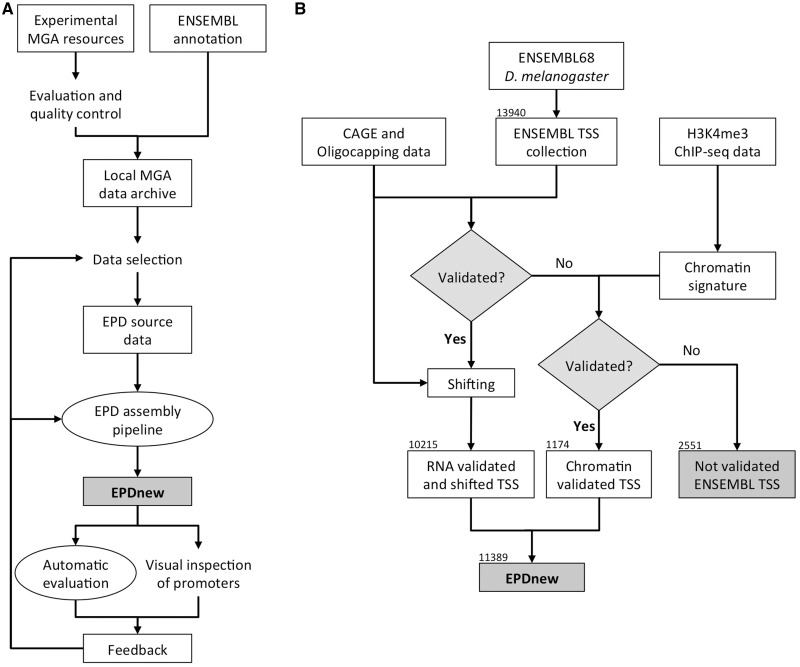
EPDnew relies on four pillars: (i) an MGA data repository which holds the primary experimental data; (ii) an automatic promoter assembly pipeline, which returns best promoter positions for an input gene set and selected MGA data; (iii) an EPD viewer, which enables both users and database curators to view promoter-defining experimental evidence in the context of other genomic features; and (iv) a sequence motif-based promoter set evaluation method. The interplay between these components is visualized in Figure 2a. External NGS data and genome annotations are first imported into the MGA repository. A part of these data is then selected as source data for the automatic assembly pipeline that generates the organism-specific promoter collections. Preliminary versions of such promoter collections are then subjected to motif-based automatic evaluation. Simultaneously, a subset of randomly chosen promoter entries (typically about 100) is visually inspected by the EPD development team. Both quality control procedures provide feedback for improving the upstream parts of the database building process, in particular the selection of the source data and the choice of the computational algorithms used to infer representative TSS positions. In practice, the preparation of a new organisms-specific promoter collection involves at least 10 such learning cycles.
Figure 2.
Work and data flow in EPDnew. (A) Physical and logical connections between source data, automatic procedures and human intervention in the development and production of EPDnew. (B) Details of the promoter assembly pipeline used in the production of the current Drosophila promoter collection of EPDnew.
The MGA data repository
The MGA data repository contains functional genomics data downloaded from public repositories such as GEO (18) along with appropriate documentation. The data are stored in a concise working format. As this resource serves other purposes, its content is not restricted to data that are of potential use for EPD. In particular, it holds a large collection of over 2000 ChIP-Seq datasets which can be explored via the ChIP-Seq server, another resource maintained by our group (see below). The datasets used by the EPD assembly pipeline are converted into genome browser viewable custom track files. In addition to primary experimental data, the MGA repository also contains manually curated genome annotation imported from other resources, for instance a compressed version of PhastCons conservation scores downloaded from the UCSC genome browser database (19).
The import of an NGS dataset into the MGA repository involves quality control steps and choices regarding data processing and data representation. In the productive format, we keep only the end positions of the mapped sequence tag. If only sequences are available from the external sources, the read mapping has to be done locally; otherwise sequence tag positions mapped by the authors are directly imported. Note further that some datasets were propagated from an earlier genome assembly to a newer one, e.g. from the mouse assembly of February 2006 (NCBI36/mm8) to July 2007 (NCBI37/mm9). The MGA data are organized as series roughly equivalent to the series in GEO (18). The procedures used to import and reformat source data into productive formats are detailed in a text document provided for each series.
EPD assembly pipeline
The EPD assembly pipeline is the central software component of EPDnew. It consists of a collection of Perl and R scripts which outputs representative promoter positions using an ENSEMBL-derived gene list and selected MGA datasets as input. The procedure differs somewhat from organism to organism. We will choose Drosophila as example for a detailed description of a promoter assembly pipeline (Figure 2b).
The procedure takes several types of inputs: (i) annotation-based input from ENSEMBL; (ii) oligo-capped 5′ tags from MachiBase (20) plus CAGE tags from (21); and (iii) ChIP-Seq data for H3K4me3 in S2 cells from (22). We used BioMart (23) to extract an ENSEMBL-based TSS collection restricted to transcripts of type ‘protein_coding’. The assembly pipeline then extracts a TSS peak list from a merged set of oligo-capping and CAGE tags. The principles of the peak-finding method were described in (5). However, we now switched to a faster software implementation based on the ChIP-Peak program (24) developed for NGS data. At the next step, the two TSS lists were compared to each other. ENSEMBL promoters with an annotated TSS falling within 50 bp of an oligo-capping/CAGE data-derived peak were selected for inclusion into EPDnew. In creating the corresponding EPDnew entries, the gene names were imported from ENSEMBL but the representative TSS position was re-defined based on the initiation site patters revealed by oligo-capping and CAGE data. The remaining non-selected ENSEMBL promoters were subsequently tested for the presence of a promoter-specific H3K4me3 signature using a novel chromatin signature matching algorithm (see Supplementary Methods). This enabled us to rescue another 1274 promoters with low mRNA 5′-end tag coverage. Note however that the TSS positions of these promoters were directly taken from ENSEMBL. The remaining 2551 promoters not supported by either type of evidence were discarded.
Similar procedures were used for generating the promoter collections for human and mouse. A technical document providing more details of the assembly pipelines can be found at the EPD website for each species and each new release. The effects of the assembly pipeline on the distribution of TSS tags, Pol-II binding and histone H3K4me3 marks are illustrated in Figure 1C for the human collection. EPDnew shows a much sharper and higher TSS peak then the corresponding ENSEMBL collection. The picture for the selected ENSEMBL promoters prior to shifting (Figure 1B) makes clear that the sharpening of the TSS peak results at least partly from an improved resolution of the TSS positions in EPDnew, in other words not merely from selection of promoters with less dispersed initiation site patterns. Sharper peaks are also observed for the Pol-II and H3K4me3 signal for EPDnew, albeit the effects are less drastic due to intrinsically lower resolution of these data.
Note that our chromatin signature-based promoter identification method requires MNase-treated ChIP-Seq data for histone modifications. Unlike oligo-capping and CAGE tags, such data are at the moment only available for a small number of tissues, which inevitably introduce a bias in the current automatically compiled chromatin-based promoter subsets. However, we believe that this bottleneck is temporary, and that a solution to the problem will soon come from more MNase data in the future. We are also aware of the fact that exclusive reliance on the H3K4me3 modification could introduce a bias against non-CpG island promoters. We are therefore working on a new protocol taking a balanced combination of several histone modifications into account.
The EPD promoter viewer
The promoter entry viewer page was completely re-designed to allow for easy inspection of the chromatin structure and other epigenetic features in the vicinity of a promoter. Rather than re-inventing the wheel, we decided to rely on the UCSC genome browser (19) as primary visualization platform. The viewer page uploads a locally stored image (originally generated at UCSC) and provides a hyperlink to UCSC enabling the user to view the same tracks directly in a browser window. The user can then further customize the track display and genome visualization range. All tracks from EPD are provided in indexed bigWig or bigBed format (25) to minimize data transfer time and volumes.
The EPD promoter viewer displays all experimental evidence that has been used for defining the reference TSS position (currently oligo-capped tags, CAGE and H3K4me3). Additional tracks are provided showing for instance the methylation status of CpG dinucleotides in the promoter region. Care is taken to represent each MGA dataset in an optional fashion. For instance, different bin sizes were used to convert different ChIP-seq datasets into wiggle files, taking into account their variation in tag density in promoter regions. To visualize the nucleosome architecture of promoters, we exclusively selected ChIP-Seq data generated with MNase digestion rather than sonication, as only the former type of data achieves single-nucleosome resolution.
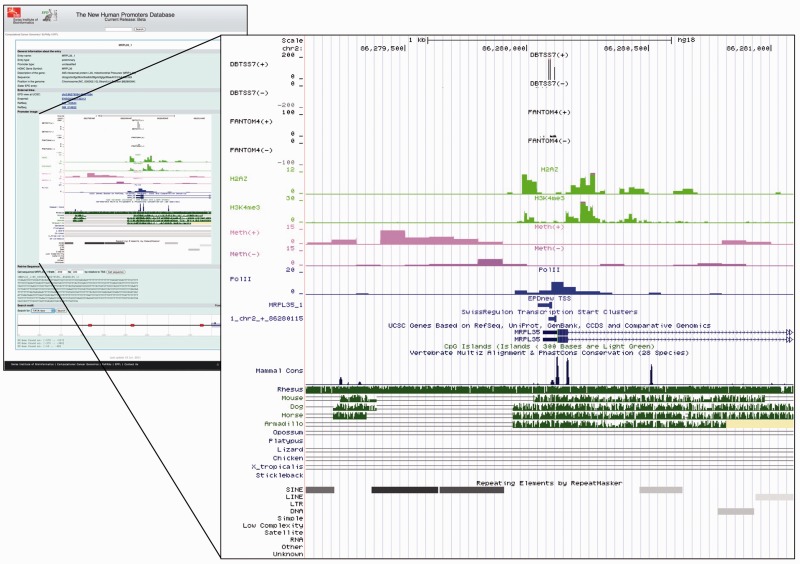
Figure 3 shows an example of a human promoter view. The TSS tracks, which are provided at single basepair resolution, are based on DBTSS version 7 (14) and FANTOM4 (26). They reflect a wide variety of tissues and cell types. ChIP-Seq data for promoter-specific histone marks and Pol-II data were taken from (7) and reflect the chromatin state in CD4+ T cells. The DNA methylome data were taken from a study carried out with IMR90 cells (8). Additional tracks from the UCSC genome browser are also included in the picture, for instance the CpG island track.
Figure 3.
EPD viewer screenshot for the human MRP-L35 promoter. The image was automatically generated and downloaded from the UCSC genome browser (15). The EPD-supplied tracks show experimental TSS sites from DBTSS7 (14) and FANTOM4 (26), chromatin marks from (7) and DNA methylome data from (8). The CpG island, genome conservation and repetitive element tracks are from the UCSC genome browser database (19). The EPD viewer page contains a link which enables users to automatically upload the EPD-supplied tracks to the UCSC genome browser for further customization and dynamic exploration of the promoter regions.
Note that the individual tracks in Figure 3 are from different cell types. The current EPD viewer thus displays a composite picture of a human promoter integrating features that may not be simultaneously present in any given cell type. As a possible future extension, we envisage to provide cell-type specific views based on data from ENCODE (27) and other epigenomics initiatives.
The MRP-L35 promoter selected for illustration exhibits average properties of a human promoter. For instance, we note a moderately dispersed pattern of TSS, four positioned nucleosomes (one upstream and three downstream of the TSS) and the usual accumulation of Pol-II signal in the internucleosomal region. Interestingly, a weak increase in unmethylated CpG (dark pink track) can be seen in the central promoter area, despite the absence of an annotated CpG island in this region.
Quality control
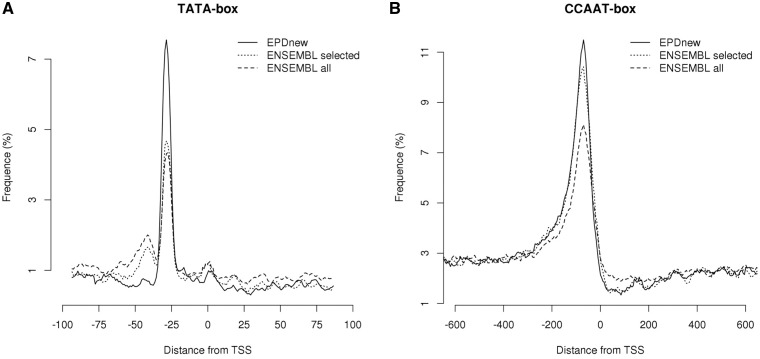
For automatic evaluation of promoter collections, we use a previously introduced benchmarking protocol based on the occurrence profiles of promoter-specific sequence motifs such as the TATA- and CCAAT-boxes (5). This protocol simultaneously measures the average precision of TSS mapping and the overall enrichment in true promoters of a promoter set. It exploits the fact that certain DNA motifs preferentially occur at characteristic distances from a TSS (28). For instance, the TATA-box occurs in a narrow region centered about 28 bp upstream of the TSS whereas the CCAAT-box occurs in a much wider area with a peak frequency at position −80. Based on these observations, we would expect a high-quality promoter collection to show high peaks for both sequence motifs. In addition, a narrow TATA-box peak at −28 would indicate precise TSS mapping.
Figure 4 shows TATA-box and CCAAT-box motif occurrence profiles for an ENSEMBL-derived human promoter collection and EPDnew. Indeed, we see higher motif occurrence peaks for the EPDnew collection. To understand whether this results from promoter selection or TSS shifting, we also show the motif occurrence profiles for the ENSEMBL promoters which were selected for inclusion in EPDnew before shifting. For the TATA-box, the increase in peak height appears to result mostly from TSS shifting, for the CCAAT-box mostly from selection. This (together with TSS profiles shown in Figure 1) suggests that EPDnew constitutes an improvement over ENSEMBL both in terms of promoter enrichment and in terms of precision in TSS assignment. Note in this context also the secondary TATA-box peak at about −40 in the ENSEMBL promoter collection, which indicates that a substantial fraction of the gene starts in ENSEMBL are in fact located 10–20 bp downstream of the true TSS. Interestingly, similar secondary peaks are seen in ENSEMBL-derived promoter collections for mouse and Drosophila, suggesting that these secondary peaks may result from an artifact of commonly used cDNA cloning techniques.
Figure 4.
DNA motif-based evaluation of promoter collections. Shown are the positional distributions of the (A) TATA- and (B) CCAAT-boxes in an ENSEMBL-derived human promoter collection, in the subset of ENSEMBL promoters that was selected for inclusion in EPD, and in EPDnew. The higher peaks and the lower back-ground frequency of the TATA-box motif in panel A indicate that EPDnew is of higher quality than ENSEMBL.
Since promoter-associated DNA motifs tend to be confined to subsets of promoters typically not exceeding 20%, we use up to eight motifs for automatic evaluation of mammalian promoter collections (Supplementary. Figure S1). Because prominent promoter motifs differ between distant taxonomic groups, we use a different motif collection for Drosophila (Supplementary Figure S2) partly based on a study by Ohler et al. (29).
We also evaluated the performance of the automatic TSS assembly pipeline by analysing the confirmation rate of promoters present in the old EPD (Supplementary Table S1). The overall confirmation rate is about 60% for human and 80% for Drosophila promoters. These rates, which may appear low, reflect the stringent validation criteria applied. With increasing data volumes entering the TSS assembly pipelines, we expect these numbers to increase rapidly in future versions of EPDnew. Interestingly, we see a higher confirmation rate for promoters of the classes ‘multiple sites’ and ‘regions’ as compared to the class ‘single initiation site’. This is consistent with the assumption that promoters with dispersed initiation site patterns were under-sampled in early versions of EPD.
We would like to stress that our DNA sequence-based evaluation method is completely independent of the experimental data used by the EPD assembly pipeline. The mRNA 5′-end mapping and chromatin profiling assays used for promoter inference are DNA sequence-blind experimental techniques. This independence of learning and evaluation data allows for iterative improvement of the promoter collections of EPDnew in a non-circular manner. Note in this context that the apparent improvement of EPDnew over ENSEMBL shown in Figure 1 would be ‘circular’ in our terminology, as we used the same data to focus the peaks. However, the concomitant sharpening and increase of the motif peak height seen in Figure 4 constitutes truly independent evidence for improvement. For this reason, we do not plan to use sequence-derived information in future promoter assembly pipelines. EPD will remain a purely experimental data-based promoter resource.
Role of human expertise
The high-quality of EPD is broadly recognized and has been attributed to manual curation by biological experts. At first sight, the transition to automatic compilation seems to call into question this advantage. This is largely a misconception. Expertise knowledge and manual curation continue to play a major role in the development and maintenance of EPDnew as well. However, these efforts are no longer targeted at individual promoter entries. The critical evaluation of experimental data takes now place at the incoming end of the database production process. Each dataset entering the MGA data repository is subjected to rigorous quality controls. A second round of evaluation is performed before a dataset is selected as input to the automatic promoter assembly pipeline. Tools offered from the ChIP-Seq server (see below) are extensively used for visual inspections of source data. For instance, plots of the kind shown in Figure 1 serve to assess the basepair resolution of ChIP-Seq data for chromatin marks. Even though we no longer check every promoter entry by eye, we continue to spend considerable time on visual inspection of randomly chosen promoter entries from EPDnew with the aid of the new entry viewer. This enables us to detect shortcomings or pathological behaviors of the automatic promoter assembly pipeline, and to amend the responsible procedures before a new promoter collection is publicly released. Human intuition-guided judgments continue to play a decisive role in this process. For all these reasons, we are confident that we will be able to maintain the quality standards of the old EPD database in the organism-specific promoter collections of EPDnew.
Coverage and comparison with other resources
The first public (and current) versions of EPDnew for human, mouse and Drosophila contain 9716, 9773 and 11 389 promoters, respectively, which is about 5 times more than the old EPD database but still not very close to complete coverage. In terms of gene coverage, we are at about 40% for human and mouse, and close to 70% for Drosophila (The new version 2 for human, which is in preparation, will contain about 15 000 promoters.). We expect to reach 90% coverage for all three species in <1 year, in view of the ongoing burst of high-throughput genomics data that are potentially exploitable for automatic promoter inference.
We used the sequence motif-based quality evaluation procedure to compare EPDnew with other publicly available human TSS collections, in particular ENSEMBL, a promoter collection provided by the FANTOM consortium, and the transcription start regions from the SwissRegulon database (30). The latter two were derived from the same CAGE dataset but using different TSS clustering strategies. Motif occurrence profiles for four prominent promoter motifs (TATA-box, CCAAT-box, GC-box and an Ets-like motif) are shown in Supplementary Figure S3. As expected, the height of the motif peaks negatively correlates with the size of the promoter collections. An overall quality ranking is impossible because each of the four collections represents a different trade-off between motif enrichment and promoter coverage. Note however that all three collections which are partly or exclusively based on CAGE data show a mono-modal TATA-box peak whereas ENSEMBL shows a multi-modal distribution, suggesting that many gene starts annotated in ENSEMBL do not correspond to a true TSS.
EPD accessory resources
The promoter collections of EPDnew can be analysed with other web services maintained by the EPD team. The Signal Search Analysis (SSA) server (31) offers tools for the discovery and characterization of DNA sequence motifs in promoter regions. The ChIP-Seq server enables EPD users to inspect the chromatin context of selected promoters, using a large collection of ChIP-Seq data from the MGA repository. Both resources are also heavily used by the EPD team in the production process of a new promoter collection. Note for instance that Figure 1 was generated with the ChIP-Cor program from the ChIP-Seq server, and Figure 4 was produced with the OProf program from the SSA server.
ACCESS
EPD (including EPDnew) is freely available without need for pre-registration. Online access is provided via the EPD web site at http://epd.vital-it.ch/. The main page contains an input text area allowing for a basic character-string search. The standard query page accepts gene symbols, and various database identifiers, in addition to free text as search criteria. It further allows for bulk download of multiple promoter sequences upon uploading a list of ENSEMBL or RefSeq gene identifiers. The latter is useful to users who would like to search promoters of differentially expressed genes identified by a micro-array experiments for over-represented sequence motifs by tools like MEME (32). The EPD and EPDnew promoter sets are also installed at the back-end of the EPD accessory servers. For instance, the ChIP-Seq server at http://ccg.vital-it.ch/chipseq/ enables users to upload their own ChIP-Seq data and analyse the intensity of the ChIP-Seq signal in the vicinity of EPD promoters by plots of the kind shown in Figure 1. Likewise, the SSA server at http://ccg.vital-it.ch/ssa/ may be used to explore the distribution of user-supplied sequence motifs.
All components of EPD can also be downloaded via FTP from ftp://ccg.vital-it.ch/ including the NGS data files stored in the MGA data repository. The old EPD promoter collections, and the organism-specific promoter collections of EPDnew are provided in various formats [for details, see (33)], including files that contain DNA sequences within a specific range relative to the TSS.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–3 and Supplementary Methods.
FUNDING
Swiss Government; Swiss National Science Foundation [31003A_125193 to G.A.]. Funding for open access charge: Swiss Government.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cavin Perier R, Junier T, Bucher P. The Eukaryotic Promoter Database EPD. Nucleic Acids Res. 1998;26:353–357. doi: 10.1093/nar/26.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki Y, Yamashita R, Nakai K, Sugano S. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tateno Y, Saitou N, Okubo K, Sugawara H, Gojobori T. DDBJ in collaboration with mass-sequencing teams on annotation. Nucleic Acids Res. 2005;33:D25–D28. doi: 10.1093/nar/gki020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid CD, Praz V, Delorenzi M, Perier R, Bucher P. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid CD, Sengstag T, Bucher P, Delorenzi M. MADAP, a flexible clustering tool for the interpretation of one-dimensional genome annotation data. Nucleic Acids Res. 2007;35:W201–W205. doi: 10.1093/nar/gkm343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J. Med. Genet. 2011;48:721–730. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 10.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 11.Rach EA, Winter DR, Benjamin AM, Corcoran DL, Ni T, Zhu J, Ohler U. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 2011;7:e1001274. doi: 10.1371/journal.pgen.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid CD, Bucher P. ChIP-Seq data reveal nucleosome architecture of human promoters. Cell. 2007;131:831–832. doi: 10.1016/j.cell.2007.11.017. author reply 832–833. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita R, Sugano S, Suzuki Y, Nakai K. DBTSS: DataBase of Transcriptional Start Sites progress report in 2012. Nucleic Acids Res. 2012;40:D150–D154. doi: 10.1093/nar/gkr1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn RM, Haussler D, Kent WJ. The UCSC genome browser and associated tools. Brief. Bioinform. 2012 doi: 10.1093/bib/bbs038. October 31 (doi: 10.1093/bib/bbs038; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahsan B, Saito TL, Hashimoto S, Muramatsu K, Tsuda M, Sasaki A, Matsushima K, Aigaki T, Morishita S. MachiBase: a Drosophila melanogaster 5′-end mRNA transcription database. Nucleic Acids Res. 2009;37:D49–D53. doi: 10.1093/nar/gkn694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, Lassmann T, Yu C, Booth BW, Zhang D, Wan KH, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, Chen X. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 2010;11:R42. doi: 10.1186/gb-2010-11-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid CD, Bucher P. MER41 repeat sequences contain inducible STAT1 binding sites. PLoS One. 2010;5:e11425. doi: 10.1371/journal.pone.0011425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skipper M, Dhand R, Campbell P. Presenting ENCODE. Nature. 2012;489:45. doi: 10.1038/489045a. [DOI] [PubMed] [Google Scholar]
- 28.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 29.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrosini G, Praz V, Jagannathan V, Bucher P. Signal search analysis server. Nucleic Acids Res. 2003;31:3618–3620. doi: 10.1093/nar/gkg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Praz V, Perier R, Bonnard C, Bucher P. The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression data. Nucleic Acids Res. 2002;30:322–324. doi: 10.1093/nar/30.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]