Abstract
Gene families often show degrees of differences in terms of exon–intron structures depending on their distinct evolutionary histories. Comparative analysis of gene structures is important for understanding their evolutionary and functional relationships within plant species. Here, we present a comparative genomics database named PIECE (http://wheat.pw.usda.gov/piece) for Plant Intron and Exon Comparison and Evolution studies. The database contains all the annotated genes extracted from 25 sequenced plant genomes. These genes were classified based on Pfam motifs. Phylogenetic trees were pre-constructed for each gene category. PIECE provides a user-friendly interface for different types of searches and a graphical viewer for displaying a gene structure pattern diagram linked to the resulting bootstrapped dendrogram for each gene family. The gene structure evolution of orthologous gene groups was determined using the GLOOME, Exalign and GECA software programs that can be accessed within the database. PIECE also provides a web server version of the software, GSDraw, for drawing schematic diagrams of gene structures. PIECE is a powerful tool for comparing gene sequences and provides valuable insights into the evolution of gene structure in plant genomes.
INTRODUCTION
In eukaryotes, a typical gene structure contains two elements: the exon and the intron. Exons are the DNA sequences that are transcribed and represented in the mature forms of RNA (mRNAs) that serve as template for synthesis of the encoded proteins. Introns that interrupt the exons in gene sequences are also transcribed, but they are removed from the mature RNA transcript by RNA splicing. Comparative analysis of exon–intron organization is important for understanding rules of gene structure and organization, protein functionality and evolutionary changes among species. The structural information of genes and gene families can serve as material for phylogenetic analyses to understand the gain, loss and change of gene structures (1–3), thereby elucidating mechanisms underlying the molecular evolution of genes and genomes (4–6). The increasing availability of plant genome sequences now makes it possible to conduct phylogenetic analyses of genes or gene families from a large number of plant species representing a large evolutionary distance. Typically, phylogenetic analyses of genes of interest require, first, the extraction of genes with corresponding intron and exon structure information, followed by phylogenetic analyses using available software programs. Comparing gene sequences to identify evolutionarily conserved gene structures is useful for predicting the biological function of protein-coding genes of interest. Accordingly, some plant comparative genomic databases, such as PlantGDB (7), PLAZA (8) and Phytozome (9), are well known and widely used because these databases allow users to extract gene structure data including exon–intron positions, exon and intron lengths and alternative splicing. Usually, users will still need to perform further analyses on the extracted data with available software programs to gain insight regarding the evolution and function of gene structure. Databases dealing with gene structure analyses are available, but with a primary emphasis on non-plant species. CIWOG is a plant database that displays common introns within orthologs in eight plant species (10). Furthermore, in most cases, phylogenetic trees, gene structures, protein domains and exon–intron comparisons for orthologs have not yet been integrated together and therefore the related databases do not provide a comprehensive view pertinent to evolution and function of gene structure. For instance, these databases do not contain information regarding which Pfam domain in a gene family contains conserved intron sites and phases. The location of introns with exons occurs as one of three different phases; between two codons (phase 0), between the first and second nucleotides of a codon (phase 1) or between the second and third nucleotide of a codon (phase 2). Intron phases are a conservative character of eukaryotic gene structures because any phase change requires either compensatory double mutations or a more complex molecular mechanism. Therefore, the location of the introns within the same sites and phase of related genes is strong support for an evolutionary relationship. Meanwhile, it is important to understand that the evolution of gene structure is often associated with the evolutionary history and functional domains of a gene of interest.
Here, we report the development of PIECE (http://wheat.pw.usda.gov/piece), a comprehensive plant gene comparison and evolution database containing all the annotated genes described from 25 plant species with available sequenced genomes. The database includes data for 17 eudicots, 5 monocots, 2 green algae and the moss Physcomitrella patens (Supplementary Table S1). The annotated genes were extracted from each species and classified based on their Pfam motif (11). Phylogenetic trees were pre-constructed for each gene category by integrating exon–intron and protein motif information. The intron site data can be shown not only in the genomic sequence but also in protein alignment sequences. The sequence and gene structure information for each identified gene is available for online access within the PIECE website. The database contains orthologs in those species for comparative analysis and evolutionary studies of gene structure. Several gene structure analysis software tools including GLOOME (12), Exalign (13) and GECA (14) have been integrated into PIECE and can be executed for each orthologous group to display exon–intron gain, loss and conservation. PIECE also provides a web interface package, GSDraw (Gene Structure Draw Server), for drawing schematic diagrams of the structures of genes derived from other species in addition to the 25 sequenced plant species. Users can submit genomic coding DNA sequence (CDS) and transcript sequences. GSDraw uses this information to obtain the gene structure, protein motif and phylogenetic tree and outputs the results as diagrams. PIECE can provide valuable information for plant researchers for analyzing the evolution of gene structure and for elucidating the biological function of proteins. PIECE is a useful resource for the research community, particularly for the study of exon–intron evolution.
DATABASE CONSTRUCTION
Data collection
PIECE currently contains a total of 947 630 annotated genes from 25 sequenced plant species including low plants to Angiosperms (Supplementary Table S1). Genome sequences, transcript sequences, protein sequences and annotation GFF files were downloaded from Phytozome (9). Exon–intron site, length and intron phase data were extracted based on the genome annotation GFF files using an in-house Java program.
Plant gene family classification
Plant genes were grouped into different families based on their protein domains using the Pfam database (v26.0) (11). We applied the hmm search program in the HMMER package (15) to search against the protein sequences of each species to classify genes. An E-value 0.01 as a cutoff, which has been widely adopted for HMMER searches, was used for queries. Many genes have more than one Pfam domain. For example, the B3 domain (PF02362) is present in either the ABI3-VP1 family or the RAV subfamily of the AP2 family. In this case, we therefore assigned PF02362 as a gene family entry that includes genes in the ABI3-VP1 and AP2 families.
Multiple sequence alignment and phylogenetic analysis
Multiple sequence alignment (MSA) was performed using the MUSCLE v3.831 program (16). The default parameters were used if the number of members in a gene family was ≤500, otherwise the option ‘–maxiters 2’ was applied. For phylogenetic analyses, the FastTree v2.1.4 program was used (17), which implements a fast and accurate approximate maximum likelihood method. FastTree analyses were conducted with default parameters; specifically, the amino acid substitution matrix used was JTT, the number of rate categories of sites (CAT model) was 20, the local support values of each node were computed by re-sampling the site likelihoods 1000 times.
Putative ortholog annotation
To predict putative orthologous relationships of genes among these plant species, we used the BLAST score ratio (BSR) method, which has been widely adopted by ENSEMBL and other studies (18). An all-against-all BLASTP search with a strict cutoff E-value <1e−20 was performed, and the BSR value was calculated for each hit. After comparing results at different BSR values, we chose a BSR value ≥0.4 as the cutoff and retrieved the top sequences in the species with the largest BSR values as the putative ortholog(s).
Orthologous gene structure evolution
PIECE uses GLOOME, Exalign and GECA for the orthologous gene structure evolution analysis. GLOOME can analyze the presence and absence profiles (phyletic patterns), which are widely used in biology (12). The default parameter settings were used for GLOOME analyses. Because the required input is a phyletic pattern provided as a 0/1 MSA, we first used MUSCLE to obtain the alignment of orthologous protein sequences with default parameters, and then calculated all intron sites. For each gene, if it has an intron site in the aligned consensus sequence, we marked ‘1’ for the site, if not, we marked ‘0’ for the site. To obtain orthologous gene exon–intron gain and loss information, a Java pipeline was implemented to include the steps described above, i.e. gene family classification, ortholog annotation and orthologous gene structure analysis.
We performed alignments between intron/exon structures using the Exalign algorithm (13). The algorithm was run in global alignment mode, and allowed intron gain/loss detection to exclude false assignments because of intron gain/loss events between orthologs. We created the Exalign dataset for each plant species in our database, and compared the full set of plant gene structures. The length of partially coding exons was adjusted to include only the coding portions. Fully non-coding exons were excluded from the comparison.
Recently, a new tool named GECA was developed, which displays gene exon–intron organization by highlighting changes in gene structure among members of a gene family (14). In PIECE, orthologs can also be displayed using the GECA method with default settings.
UTILITY AND WEB INTERFACE
PIECE is a web-based tool combining a MySQL database management system with a dynamic web interface based on PHP and Javascript. The exon–intron data in the database are searchable and viewable.
Search system
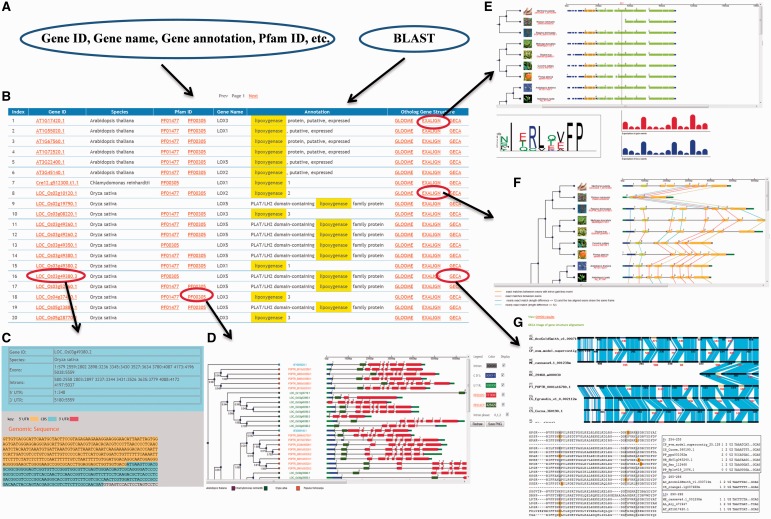
PIECE has a user-friendly entry point for searching each gene. Users can retrieve any gene by a keyword search for gene ID, gene name or gene function or by a BLAST search using either the nucleotide or protein sequence of your gene of interest (Figure 1A). The main page of the search results lists all genes meeting the search criteria and provides brief information, such as gene accession, gene description, source organism, gene annotation, Pfam domain and gene ortholog analysis (Figure 1B). The columns in Gene ID, Pfam ID and Ortholog Gene Structures are linked to more detailed information of the analysis results. For example, clicking on a gene accession will display details on the exon–intron information for each gene, including the genomic and transcript sequences (Figure 1C). Each gene in the search result contains the location of the Pfam domain that was identified in its protein sequence. Clicking on a Pfam domain will show its phylogenetic tree along with gene structures (Figure 1D). The ortholog analysis link will display the GLOOME, Exalign and GECA results of the gene (Figure 1E and F), which link to details that include more elaborate descriptions of the orthologous gene structure evolution results. The detail of the gene structure and evolution analysis in the Pfam ID and Ortholog Gene Structures columns are also presented (see below).
Figure 1.
Graphical user interface overview. The user interface provides multiple ways to access information stored in the database. (A) Multiple inputs for PIECE. (B) Search results interface. (C) Sequence detail including exon–intron information. (D) The graphical viewer for PIECE. (E) GLOOME analysis results. (F) Exalign analysis results. (G) GECA (CIWOG) analysis results.
Phylogenetic tree display along with gene structure and Pfam domains
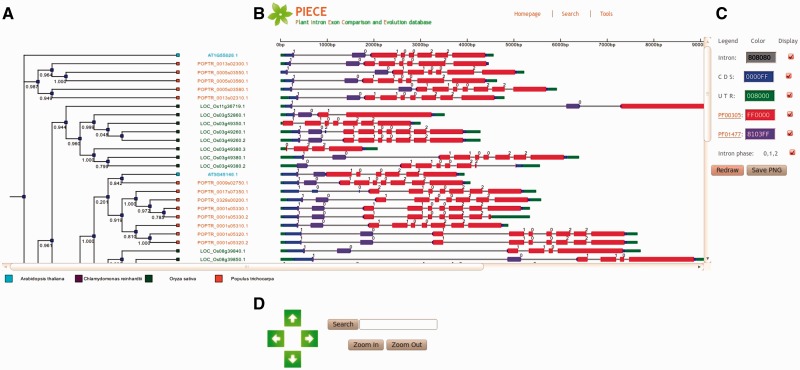
The database provides a user-friendly graphical view that displays SVG-formatted output, which contains a gene structure and Pfam domain pattern diagram linked to a bootstrapped similarity dendrogram (Figure 2). Depending on the annotations present in the database, the viewer can automatically recognize elements of the gene structure, such as coding exons, introns and UTRs. Default conventions are used to render exons (thick boxes), UTRs (thin blue and green boxes) and introns (thin grey boxes), but the user can modify the display of the elements by selecting a different color or choosing to not display the element. A search function is provided to allow users to search the gene ID in he phylogenetic tree. If the ID is found, it will be highlighted in red. Controls available on the bottom of the page allow magnification of tree image (e.g. the ‘zoom in’ and ‘zoom out’ buttons) as well as movement of the magnified image with the arrow buttons. When viewing the gene structure, the exons, introns and Pfam domains for genes can be selected easily. When the user hovers the mouse over each element, the length of the element will be shown. By clicking the element, the sequence information for the selected element will be displayed. As a demonstration, the analysis results for the Lipoxygenase gene family are presented in Figure 2.
Figure 2.
The PIECE viewer. Data for the LOX gene family (PF00305) in Arabidopsis thaliana, rice, poplar and Chlamydomonas reinhardtii. (A) Dendrogram of sequences clustered according to the presence and similarity of extracted Pfam motifs. (B) Diagram that displays positional information of the gene structure in each sequence. (C) Color selector and check boxes for displaying introns, CDS, UTRs, Pfam domains and intron phases, and save button to save the output as a PNG file. (D) Color for the plant species and operation panel for manipulating the output.
Multiple types of gene structure display
Gene structure visualization is important for analyzing exon–intron evolution. Typically, the basic components of gene structure (UTRs, intron, exons) are displayed on genomic sequence (19,20). To find relationships between exon–intron compositions in the encoded proteins, exon boundaries are also mapped onto the protein sequence (21,22). The view function in PIECE provides three types of exon–intron displays for each Pfam domain. Users can select any protein domain of interest by clicking the Pfam ID in the search results.
Analysis of gene structure evolution with groups of orthologs
On a gene family scale, global analysis is useful for dating intron changes; however, for certain genes, gene structure evolution in different species is not clear in phylogenetic trees with exon–intron pattern diagrams. Moreover, not all genes contain Pfam motifs in their encoding proteins and therefore cannot be analyzed as in Figure 2. Consequently, it is necessary to show exon–intron fluctuations for each gene in the database because intron-containing genes are spread across diverse plant phyla, whereas orthologs often have similar exon–intron organization even at large evolutionary distances (23). PIECE provides three analysis methods for each gene to infer the evolution of exon–intron structure in multiple protein-coding ortholog sets along a fixed-species phylogeny.
GLOOME
To analyze the gain and loss of introns in the ortholog group, we used GLOOME, which accurately infers branch-specific and site-specific gain and loss events with presence and absence profiles. To integrate GLOOME into PIECE, we first aligned the protein sequences within the ortholog group. We next coded intron characteristics using binary characters to denote presence (‘1’) and absence (‘0’). The 0/1 matrix, in which rows correspond to species and columns corresponds to binary characters, is termed a phylogenetic profile of presence–absence or phyletic pattern and is equivalent to a MSA. In PIECE, the output of GLOOME includes plant species trees, gene structure displays, intron site sequence logos and the expected number of gains and losses for each intron site (Figure 1E). When users click each box in the histogram, the viewer will show the intron site in the aligned protein sequence. The alignments were generated using MUSCLE, and the sequence alignment graphical display was implemented in the Jalview (24) Java applet. GLOOME provides useful analytical facilities for exploring the degree of conservation of intron evolution across proteins in the ortholog group and also for analyzing the distribution of exonic sequences within the aligned coding sequences of domains.
Exalign
During evolution, one exon may split into multiple exons or multiple exons may fuse into one; such events have stringent constraints in exon length, and this characteristic can be used to determine cases of exon fusion or division. To analyze the evolution of gene structure of orthologs, we use another tool named Exalign (13). The Exalign viewer of PIECE can show the relationship of exons in orthologous genes from different plant species. This viewer provides exon–intron display for orthologs of gene structure data sets linked to the species phylogeny (Figure 1F). The gene-exon comparison between species is shown as colored lines. Different colors indicate different exon comparison results. In PIECE, any gene data with its set of orthologs can be put into the Exalign viewer at the user’s request to easily find the evolution history of genes and, particularly, to detect exon relationships and fusion events.
GECA
Aligning exon–intron structures accompanied with similarities between sequences is helpful for annotating gene structure information. GECA can analyze gene exon–intron organization and highlight changes in gene structure (14). GECA relies on protein alignments, completed with the identification of common introns in corresponding genes using CIWOG (Common Introns Within Orthologous Genes) (10). In PIECE, each gene has a GECA link to view the orthologs that are aligned using their common introns detected by CIWOG. The similarities between orthologous sequences in the alignment are represented at the level of amino acids in the translated exons. A blue line links two amino acids if they are identical, a purple line indicates conservative substitutions, and intron type is detected by CIWOG (Figure 1G).
GSDraw web server
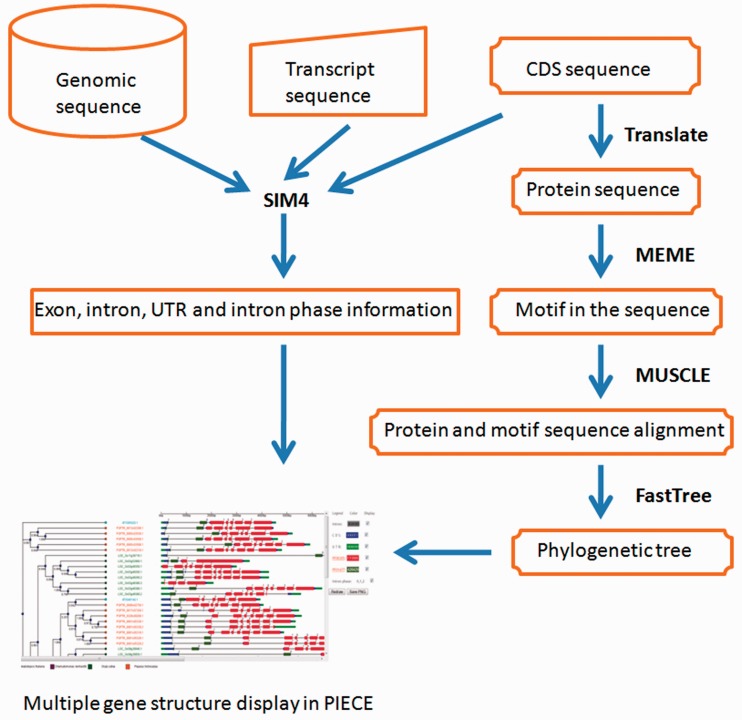
A number of web tools have been developed for gene structure annotation, such as GSDS (19), FancyGene (20) and GECA (14). The purpose of these programs is to represent the exon–intron structure of several genes in a single image to perform global gene structure comparisons (14). However, these resources do not display sequences with phylogenetic relationships and automatically detected protein motifs. Therefore, we developed GSDraw as part of PIECE. GSDraw is a convenient and easy-to-use interface for gene structure annotation that integrates Sim4 (25), MEME (26), MUSCLE (16) and FastTree (17) into a single web-based tool. The procedures for designing and implementing the GSDraw server are illustrated in Figure 3. Users submit a query sequence set (in multi-FASTA format) consisting of genomic, CDS or transcript sequences to GSDraw (http://wheat.pw.usda.gov/piece/GSDraw.php) and obtain schematic diagrams of their gene structures with annotated Pfam protein motifs and a phylogenetic tree. This capability allows users to view a PIECE database-style display for any selected gene family group (of three or more genes) from any species with available data. The GSDraw output for three rice LRR-Kinase genes is shown in Supplementary Figure S1. The user can modify the gene structure display to their own preferences by selecting different colors for the annotated sequences and/or choose whether or not to display each of the Pfam motifs, similar to what is allowed in the PIECE viewer.
Figure 3.
Workflow chart of GSDraw.
DISCUSSION
Simple sequence alignment and comparison usually is unable to provide a clear picture of the structural evolution of genes, e.g. how their intron–exon structures, intron lengths, alternative splicing and untranslated regions change over time. Although there has been a rapid growth in the number of plant genome databases, such as PlantGDB (7), PLAZA (8), Phytozome (9) and GreenPhylDB (27), these resources lack comparative analytical capabilities for integrating protein domains from multiple species to investigate exon–intron structural evolution. ExDom (28) contains an extensive collection of exon–intron gene structures mapped to protein domains, but it primarily focuses on non-plant species. Furthermore, most related databases do not display the phylogenetic tree of gene families and orthologous gene evolution histories. To address these limitations, we developed PIECE, which characterizes the number, position and length of introns and exons from 25 individual sequenced plant genomes. The PIECE database provides a panoramic perspective from which to investigate the evolution of gene structures on a broad evolutionary time scale. Furthermore, PIECE provides an easy entry point for researchers to immediately access gene structure evolution information without having to install any software.
For example, heat shock response in eukaryotes is transcriptionally regulated by conserved heat shock transcription factors (Hsfs). Hsf genes are represented by a large multigene family in plants. To illustrate the possible mechanisms of structural evolution of Hsf homologs, we used PIECE to compare the exon–intron structures of individual Hsf genes in 10 plant lineages. Supplementary Figure S2 provides a detailed illustration of the relative length of introns and the conservation of the corresponding exon sequences within each of the Hsf genes. Notably, although the members of the Hsf gene family exhibited differences in intron number and intron length, the intron positions and intron phases were remarkably well conserved, with conserved splicing sites between adjacent exons.
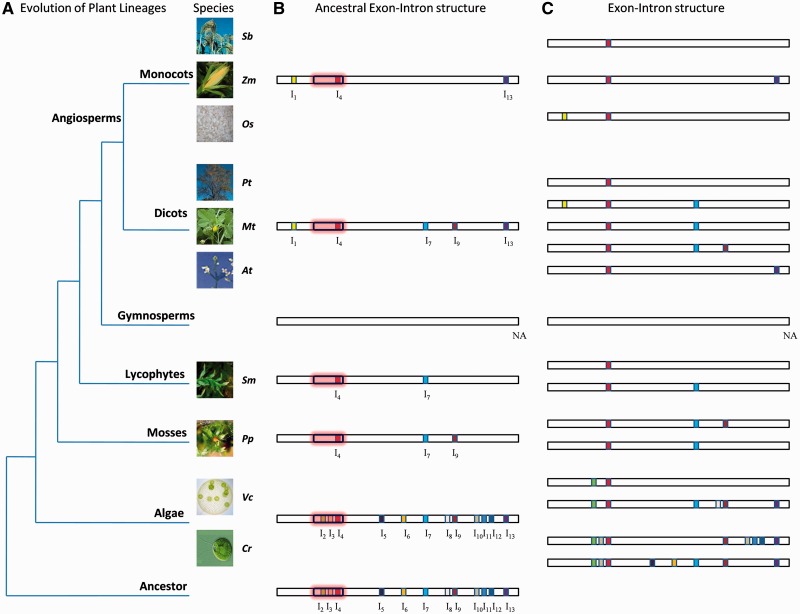
To further investigate the structural evolution of Hsf genes in different lineage species, we also used PIECE to create images that contain gene structure information in unaligned and aligned protein sequences (Supplementary Figures S3 and S4). We next constructed an evolutionary model that could predict the current Hsf genes in plant species of different lineages (Figure 4). Under the assumption that introns, which were located at identical positions and given identical phase, should be present in the common ancestor, we reconstructed the ancestral exon–intron structure of Hsf for all plant lineages (Figures 4). The results obtained from the Hsf intron analysis suggested that the ancestral Hsf contained ≥12 introns, symmetrically distributed throughout its coding sequence (Figure 4). The aquatic plants (green algae) have a large number of introns. Most introns were lost in the evolution of aquatic plants (green algae) to lower land plants (mosses and lycophytes), including I2, I3, I5, I6, I8 and I10–I12. Moreover, single intron losses also occurred during the expansion and divergence of the Hsf gene family in each plant lineage. For example, the ancestral Hsf in monocots contained at least 3 introns, whereas all Hsf genes in monocots contained only 1 or 2 introns (Figure 4B and C). It appears that I7 and I9 are not present in monocots, but are present in the dicot ancestor. Besides the intron loss, gain of an intron is also observed. I1 is only present in angiosperms. Furthermore, the analysis revealed that I4 is present in the Hsf gene of the common ancestor of all plant lineages (Figure 4B), and its position is in the DNA binding domain (DBD) (Figure 4C). This observation indicates that the Hsf family in plants not only has a conserved DBD motif but also contains a conserved intron in the DBD domain.
Figure 4.
An evolutionary model for the structural evolution of the Hsf gene family in plants. (A) Dendogram representing the evolutionary relationship of all plant lineages. (B) Proposed exon–intron structure of the ancestral Hsf gene in each plant lineage. (C) Current exon–intron structure of Hsf genes. The exon–intron structure of the Hsf genes in gymnosperms is represented with a empty bar because genomic sequences are unavailable.
As the examples demonstrate, the capabilities of PIECE will provide researchers with many hypotheses for designing molecular biology studies and will help to elucidate the evolutionary history of plant genes. Future efforts will extend the number of available plant species and enhance the analytical capabilities of PIECE. Newly published plant genomes will enable efficient phylogenetic analyses of exon-domain relationships in plants and in-depth analysis of the evolutionary history of protein domains. Alternative splicing is an important biological process that greatly increases the biodiversity of proteins that can be encoded by the genome. One of the future directions will focus on the integration of alternative splice data into PIECE for gene evolution and structure analyses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
U.S. National Science Foundation [IOS 0701916 and IOS 0822100]; United State Department of Agriculture, Agriculture Research Service CRIS project [5325-21000-014]. Funding for open access charge: US Department of Agriculture, Agriculture Research Service.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Eric Yan for his assistance on the database development and Xiaohao He for the critical reading of the manuscript. The authors are very grateful to Farhad Ghavami, Ajay Kumar, Vijay Tiwari, Jiajie Wu, Filippo Bassi and the anonymous reviewers for testing the server and offering valuable comments.
REFERENCES
- 1.Javelle M, Klein-Cosson C, Vernoud V, Boltz V, Maher C, Timmermans M, Depege-Fargeix N, Rogowsky PM. Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiol. 2011;157:790–803. doi: 10.1104/pp.111.182147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turchetto-Zolet AC, Maraschin FS, de Morais GL, Cagliari A, Andrade CM, Margis-Pinheiro M, Margis R. Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evol. Biol. 2011;11:263. doi: 10.1186/1471-2148-11-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strommer J. The plant ADH gene family. Plant J. 2011;66:128–142. doi: 10.1111/j.1365-313X.2010.04458.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Z, Zhang Y, Long M. Extensive structural renovation of retrogenes in the evolution of the Populus genome. Plant Physiol. 2009;151:1943–1951. doi: 10.1104/pp.109.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Espana A, Mares R, Sun TT, Desalle R. Intron evolution: testing hypotheses of intron evolution using the phylogenomics of tetraspanins. PLoS One. 2009;4:e4680. doi: 10.1371/journal.pone.0004680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin H, Zhu W, Silva JC, Gu X, Buell CR. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006;7:R41. doi: 10.1186/gb-2006-7-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, Lushbough C, Brendel V. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–D965. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkerson MD, Ru Y, Brendel VP. Common introns within orthologous genes: software and application to plants. Brief Bioinform. 2009;10:631–644. doi: 10.1093/bib/bbp051. [DOI] [PubMed] [Google Scholar]
- 11.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen O, Ashkenazy H, Belinky F, Huchon D, Pupko T. GLOOME: gain loss mapping engine. Bioinformatics. 2010;26:2914–2915. doi: 10.1093/bioinformatics/btq549. [DOI] [PubMed] [Google Scholar]
- 13.Pavesi G, Zambelli F, Caggese C, Pesole G. Exalign: a new method for comparative analysis of exon-intron gene structures. Nucleic Acids Res. 2008;36:e47. doi: 10.1093/nar/gkn153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fawal N, Savelli B, Dunand C, Mathe C. GECA: a fast tool for gene evolution and conservation analysis in eukaryotic protein families. Bioinformatics. 2012;28:1398–1399. doi: 10.1093/bioinformatics/bts153. [DOI] [PubMed] [Google Scholar]
- 15.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo AY, Zhu QH, Chen X, Luo JC. [GSDS: a gene structure display server] Yi Chuan. 2007;29:1023–1026. [PubMed] [Google Scholar]
- 20.Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;25:2281–2282. doi: 10.1093/bioinformatics/btp381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leslin CM, Abyzov A, Ilyin VA. Structural exon database, SEDB, mapping exon boundaries on multiple protein structures. Bioinformatics. 2004;20:1801–1803. doi: 10.1093/bioinformatics/bth150. [DOI] [PubMed] [Google Scholar]
- 22.Csuros M. Malin: maximum likelihood analysis of intron evolution in eukaryotes. Bioinformatics. 2008;24:1538–1539. doi: 10.1093/bioinformatics/btn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- 24.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouard M, Guignon V, Aluome C, Laporte MA, Droc G, Walde C, Zmasek CM, Perin C, Conte MG. GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res. 2011;39:D1095–D1102. doi: 10.1093/nar/gkq811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhasi A, Philip P, Manikandan V, Senapathy P. ExDom: an integrated database for comparative analysis of the exon-intron structures of protein domains in eukaryotes. Nucleic Acids Res. 2009;37:D703–D711. doi: 10.1093/nar/gkn746. [DOI] [PMC free article] [PubMed] [Google Scholar]