Abstract
The National Institutes of Health Genetic Testing Registry (GTR; available online at http://www.ncbi.nlm.nih.gov/gtr/) maintains comprehensive information about testing offered worldwide for disorders with a genetic basis. Information is voluntarily submitted by test providers. The database provides details of each test (e.g. its purpose, target populations, methods, what it measures, analytical validity, clinical validity, clinical utility, ordering information) and laboratory (e.g. location, contact information, certifications and licenses). Each test is assigned a stable identifier of the format GTR000000000, which is versioned when the submitter updates information. Data submitted by test providers are integrated with basic information maintained in National Center for Biotechnology Information’s databases and presented on the web and through FTP (ftp.ncbi.nih.gov/pub/GTR/_README.html).
INTRODUCTION
Technological advances have accelerated discoveries about human genetic disorders and fueled growth in both the number and complexity of genetic tests. Yet, without ready access to accurate and detailed information about tests, health care providers cannot make informed clinical decisions about the use of genetic tests for patient care.
To enhance access to test information, the National Institutes of Health (NIH) developed the Genetic Testing Registry (GTR, www.ncbi.nlm.nih.gov/gtr/), a free online resource that centralizes comprehensive information about genetic tests offered in the USA and abroad. The information is voluntarily submitted by test providers through a variety of electronic formats. The unit of record is an orderable test, each of which is assigned a unique accession number. A genetic test is defined by GTR as an analysis of human chromosomes, deoxyribonucleic acid, ribonucleic acid, genes and/or gene products (e.g. enzymes and other types of proteins), which is predominately used to detect heritable or somatic mutations, genotypes or phenotypes related to disease and health (adapted from http://oba.od.nih.gov/oba/SACGHS/reports/SACGHS_oversight_report.pdf). The Registry includes information about health-related clinical and research tests for germline variation, including pharmacogenetic tests, and soon will expand to include tests for somatic variants. Biochemical, cytogenetic and molecular genetic tests are all within the scope of GTR.
The GTR organizes critical information such as the purpose of the test, target population, test methods, what the test measures, analytical validity, clinical validity, clinical utility, ordering information and test credentials [e.g. Food and Drug Administration (FDA) approval or clearance] as well as the laboratory name, location, contact information, certifications and licenses. The website links to context-specific information about medical conditions, genes, sequence variation, test standards, practice guidelines, pharmacogenetic information, clinical trials, molecular resources and consumer support sites. The primary audience of the initial phase of GTR is the health care community.
BACKGROUND
The GTR arose from the 2008 recommendation by the Secretary’s Advisory Committee on Genetics, Health and Society (SACGHS) that called for a publicly available web-based registry to enhance the transparency of genetic testing and to assist efforts in reviewing the clinical validity of laboratory-developed tests (LDTs) (http://oba.od.nih.gov/oba/SACGHS/reports/SACGHS_oversight_report.pdf/). Other policy (1) and advocacy (2) groups also called for a registry to represent the risk continuum and enable informed decision-making regarding genetic testing.
The GTR was established at the National Center for Biotechnology Information (NCBI) under the oversight of the NIH Office of the Director. NCBI was identified as the ideal place to implement and maintain the database given its extensive experience with integrating access to information in the domains of genomic sequence and sequence variation, genotype–phenotype relationships and medical literature.
The GTR is maintained in consultation with Federal agencies—FDA, the Center for Medicare and Medicaid Services (CMS), the Centers for Disease Control and Prevention (CDC) and the Federal Trade Commission—and with extensive input from health care providers, researchers, testing laboratories, patient advocacy groups and other stakeholders. In addition, two advisory committees are consulted on an ongoing basis—the NIH Clinical Advisors to the GTR (roster at http://oba.od.nih.gov/oba/gtr/Roster_NIH_Clinical_Advisors_to_GTR_1-5-12.pdf), a group of clinical experts from within NIH, and the Medical Genetics Working Group of the Board of Scientific Counselors of the NCBI (roster at http://oba.od.nih.gov/oba/gtr/Roster_NCBI_BSC_Medical_Genetics_Working_Group_1-5-12.pdf). Information about the GTR and its governance is maintained by the NIH Office of Science Policy (http://oba.od.nih.gov/gtr/gtr.html) and is available from the GTR home page.
SCOPE OF THE DATABASE
The GTR maintains comprehensive documentation about data that may be submitted to describe registered laboratories and tests (ftp://ftp.ncbi.nlm.nih.gov/pub/GTR/documentation/GTRFieldDefinitions.pdf). Data fields are grouped into three categories: minimal (required elements for submitters who choose to register information about their laboratory and tests), recommended and optional (http://www.ncbi.nlm.nih.gov/gtr/docs/fieldrequirements/). This categorization reflects the importance of understanding the evidentiary basis for claims made about tests and the information laboratories can be expected to have based on current regulatory oversight (discussed in the following section, increasing transparency through the GTR).
The GTR is designed to standardize representation of laboratory- and test-specific information. The content provided by the submitter to GTR is integrated with information from other NCBI databases. Whenever possible, data are processed using controlled language and specific fields rather than free text. Identifiers used in other databases to represent concepts of interest to GTR’s users are maintained by GTR not only to support linking to those databases, but also to facilitate data exchange. Data managed by GTR, often in collaboration with other groups, are provided for unrestricted access through File Transfer Protocol (FTP).
Laboratories may participate in GTR if their tests are in scope. Laboratories that participate in the GeneTests Laboratory Directory may register in GTR without specific approval; new labs are reviewed by GTR staff before they are approved to register tests (http://www.ncbi.nlm.nih.gov/gtr/docs/submit/#who).
The GTR contains information about laboratories and the tests they offer but does not contain or gather information on genetic test results. Ancestry testing is not regarded as health-related and is therefore not in scope. Facilities only collecting or preparing specimens (or both) or only serving as a mailing service and not performing testing are not considered by the Clinical Laboratory Improvement Amendments of 1988 (CLIA) to be laboratories (http://wwwn.cdc.gov/clia/regs/subpart_a.aspx#493.2) and thus are not in scope for GTR.
Increasing transparency through the GTR
Although oversight of genetic tests and the laboratories performing them is provided at the Federal level by FDA and CMS, respectively, the 2008 SACGHS report highlighted several regulatory gaps and ambiguities. The NIH is not a regulatory agency and, thus, it cannot enforce Federal regulations nor mandate the submission of information to the GTR. However, consolidating information in a publicly accessible format can serve to reveal problems or questions, which may then prompt evaluation and action by other Federal agencies and/or professional organizations.
Analytical validity
In the USA, clinical tests may be provided only by laboratories certified by CMS through CLIA or by CLIA-exempt laboratories (http://wwwn.cdc.gov/clia/regs/subpart_a.aspx#493.2). For CLIA certification, laboratories undergo inspections during which they may be required to establish and verify the analytical validity (i.e. the accuracy and reliability with which a test measures the component of interest such as genotype or an analyte) of a subset of tests they offer. The inspection process is not designed to assess every test, and the analytical validity measures reported by laboratories are not collected in a CMS database. Moreover, information about analytical validity is not always readily available from US clinical laboratories. To help fill this information gap, analytical validity is a minimal field for submitters who choose to register clinical tests in the GTR.
Clinical validity
Information about clinical validity, a measure of how consistently and accurately the test detects or predicts the intermediate or final outcomes of interest, is important to clinicians making decisions about whether to order a test. Clinical validity depends on the population in which the testing is performed; thus, information about the appropriateness of the test may be limited to populations for which data exist. There are no Federal requirements for laboratories to establish or verify the clinical validity of tests that they offer. Most genetic tests are LDTs, which are solely for use within the developer’s laboratory. CMS does not have authority under CLIA to require the documentation of clinical validity. As part of its charge to evaluate safety and effectiveness, FDA has the authority to evaluate associations between genetic markers and clinical diagnosis in the context of intended use. However, FDA has exercised enforcement discretion and generally has not enforced applicable regulations with respect to LDTs. Information about clinical validity, target populations and supporting publications are recommended fields for test registration in the GTR.
Clinical utility and Purpose of the test
Tests are performed for specific clinical purposes and clinical utility, how likely the test is to significantly improve patient outcomes, is a measure of the usefulness of a test. Clinical utility information is essential for making medical management decisions, developing professional guidelines and making coverage decisions. Insufficient evidence of clinical utility for most tests was identified by SACGHS as an important gap in the oversight of genetic testing.
Laboratories typically do not have the capability to gather and publish evidence of clinical utility, but can report on the available evidence. Test submitters may choose from a standardized list of categories of purpose of the test (Table 1) and clinical utility (Table 2). Purpose of the test is a minimal field. Clinical utility is a recommended field, and for each category chosen, a supporting citation or Uniform Resource Locator (URL) must be provided.
Table 1.
Category choices for select fields with medical importance—purpose of the test
| Category choice | Definition |
|---|---|
| Diagnosis | Identification or confirmation of disease |
| Drug response | Evaluation of sequence variation influencing an individual’s reaction to specific medications, as in a pharmacogenetic test. Excludes immune-mediated adverse drug reactions (dose-independent drug allergies). |
| Monitoring | Periodic or continuous evaluation of a disease or condition over time, including a patient’s response to medical treatment. |
| Mutation confirmation | Re-evaluation of a genetic test result to assess the validity of the initial result. For example, research test results or results from another laboratory. |
| Pre-implantation genetic diagnosis | Genetic testing performed on a small number of cells from a human embryo before uterine implantation as part of assisted reproduction procedures. |
| Pre-symptomatic | Genetic analysis of an asymptomatic or unaffected individual who is at risk of a specific genetic disorder. |
| Risk assessment | Evaluation of the likelihood of developing a specific condition based on genetic risk. Includes carrier testing in affected families. |
| Screening | Evaluation of a target population to identify a subgroup affected by a genetic condition or that have the potential to transmit the trait to their offspring. Includes newborn screening, ethnicity-based screening and pre-conceptual genetic testing. |
Table 2.
Category choices for select fields with medical importance—clinical utility
| Category choice |
|---|
| Avoidance of invasive testing |
| Establish or confirm diagnosis |
| Guidance for management |
| Guidance for selecting a drug therapy and/or dose |
| Lifestyle planning |
| Predictive risk information for patient and/or family members |
| Reproductive decision-making |
| Sufficient research has not been conducted to demonstrate the utility of the test |
Published practice guidelines developed by professional organizations exist for a subset of genetic conditions and inform the evidence base for clinical utility. These guidelines are gathered by the GTR curators and displayed alongside tests and conditions using the Discovery panel (Figure 1B). This layout enables a comparison view for users where the assertions of the professional societies can complement the material gathered by the laboratory. Over time, the manual assembly of genetic practice guidelines into a centralized location—which is at present not otherwise available—will serve as a useful resource.
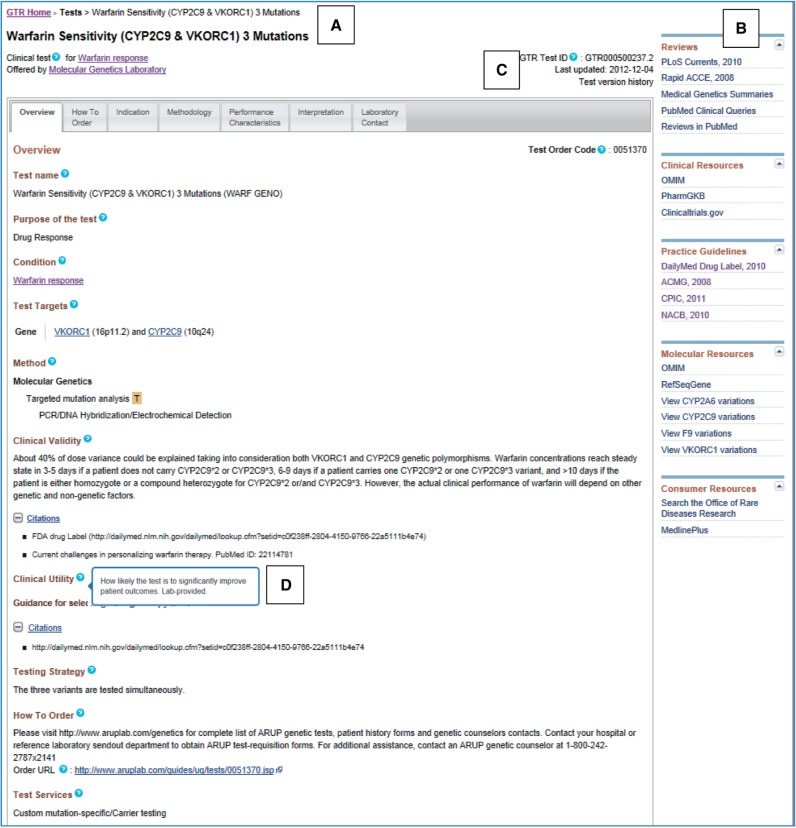
Figure 1.
Representative clinical test record displaying overview tab. Information is organized along seven tabs. (A) Breadcrumbs summarize the search path. (B) Discovery panel, located on test- and condition-specific pages, includes links of interest for the condition. (C) A unique GTR accession is assigned to each test record. The date the record was last updated and test version history are provided. (D) Tooltips, accessed by hovering over a ‘?’ icon, display the definition of specific fields and identify fields that are provided by submitters. In this example, hovering over the tooltip for the clinical utility field prompts the tooltip display.
Phenotypes and conditions
Management of information about phenotype in the GTR is founded on the Unified Medical Language System (UMLS®) (3). Unique identifiers are assigned to the concept of a condition, and alternate terms used by different sources are grouped by the same identifier. Relationships between concepts, such as hierarchies and features characteristic of a disorder, are also maintained. If a term is not represented in the UMLS, a combination of computational and curatorial approaches is used to assign the term to an existing concept, or to create a new concept. For example, general terms may be introduced to support testing related to common conditions such as pharmacogenetic responses or tests covering a broad scope of conditions. With each release of the UMLS, identifiers created by NCBI are updated to match those of UMLS as appropriate.
UMLS is the GTR’s source for condition names from Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT®), International Classification of Diseases, ninth revision (ICD-9), International Classification of Diseases, tenth revision (ICD-10) and Medical Subject Headings (MeSH) and the mapping between Mendelian Inheritance in Man (MIM) numbers and concept identifiers. When available, SNOMED CT® terms are used as the preferred term for any concept. This convention is used to enable interoperability with electronic medical records and because SNOMED CT® terms are now provided without licensing restrictions. Clinical features characteristic of a disorder are reported from the Human Phenotype Ontology (HPO) (4), and data from the Online Mendelian Inheritance in Man (OMIM) (5) are integrated into the UMLS.
A subset of alternate names, the preferred term and their symbols are used to construct a dictionary that supports interactive queries. The terms selected as preferred by the GTR are now being used in the phenotype section of NCBI’s Gene database (6) to standardize reporting among NCBI’s databases.
The GTR solicits feedback about condition names, curates records and collaborates with data sources as appropriate. This feedback loop is intended to help refine the accuracy and usability of the genetic nomenclature over time.
Genes, proteins, cytogenetic locations, variation and drug names
The GTR uses official gene symbols and full names from the Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) (7), as processed by NCBI’s Gene database. NCBI’s GeneID’s, gene-specific summaries and cytogenetic data are integrated automatically. Protein names are used from reviewed Universal Protein Resource Knowledgebase (UniProtKB) (8) records, through NCBI’s Reference Sequence collection (RefSeq) (9). Drug names, and their identifiers, are incorporated from Pharmacogenomics Knowledgebase (PharmGKB) (10).
Submitters are encouraged to provide information about the alleles they test in a form accepted by the Human Genome Variation Society (HGVS) (11), but other formats are allowed. Only variations represented by HGVS terms can be mapped to the genome reference to support browser access to tested variations. GTR encourages submitters to volunteer information about the alleles on which pathogenicity is asserted (clinical significance) to increase transparency. NCBI calculates the set of known variants in a specified genomic region, and GTR can provide information to the user about assertions of pathogenicity that have been reported elsewhere. We will continue to build views that enable users to anticipate the set of known variants that might be reported back on a test and to evaluate the report, once received. This will be increasingly important as tests broaden in the scope to include gene panels and whole exomes.
Related literature
GTR maintains context-specific records from the medical and scientific literature and also links to GeneReviews, a set of expert-authored, peer-reviewed, clinical descriptions of ∼550 inherited conditions (http://www.ncbi.nlm.nih.gov/books/NBK1116/). GeneReviews, Medical Genetic Summaries (http://www.ncbi.nlm.nih.gov/books/NBK61999/) and Clinical Pharmacogenetics Implementation Consortium guidelines (12) are excerpted for display on the website. Submitters may provide references supporting their submissions (e.g. for the clinical validity field) and in certain cases a citation must be provided if the submitter chooses to complete a recommended field (e.g. for the clinical utility field). Users may also suggest that particular published practice guidelines or scholarly publications be added to the GTR (www.ncbi.nlm.nih.gov/gtr/feedback/).
Accessioning and versioning of test records
Each test is assigned a unique GTR accession number by NCBI that can be used to retrieve the current version of that test. In addition, each accession is given a version to enable tracking history and retrieval of previous versions of a particular test. If a submitter updates a test, a new public version is assigned by NCBI and the record is refreshed. If key elements in the GTR database that affect the content of automatically provided data are modified, the public test record is updated but no new version is assigned (e.g. changes in cytogenetic locations for genes or preferred names for conditions). The version history of each test is retained in the database so that if a test is referenced in publications, the authors can refer to the specific accession and version and anyone will be able to view that version at a later date. The version history will be made available from the test-specific page.
The accessioning of well-described genetic tests facilitates formal evaluation by professionals and can be used in publications of these evaluations. NCBI has encouraged the American College of Medical Genetics and Genomics (ACMG), the American Society of Human Genetics (ASHG), the Association for Molecular Pathology (AMP) and the National Society of Genetic Counselors (NSGC) to publish evaluations of the tests in the GTR. Professional evaluations can be displayed in the GTR as with other important publications (e.g. practice guidelines) that would further help fill information gaps about genetic tests.
PROCESSING SUBMISSIONS
The GTR supports registration of tests in multiple ways. An interactive web interface is implemented as a component of NCBI’s submission portal (https://submit.ncbi.nlm.nih.gov/subs/gtr/). Users are authenticated using the MyNCBI system and authorized to submit based on having a previous account in the GeneTests Laboratory Directory (13), or approval by GTR staff. A wizard guides the submitter through the process, providing suggestions such as standard names of conditions, genes, proteins, variations and cytogenetic locations. If minimal fields are not completed by the submitter, the wizard highlights what is missing and prompts for missing data. At the end of the process, the submitter can review the submitted text and preview the results as they will be displayed on the web interface. A complete, printable guide to the submission process is also provided on the web (www.ncbi.nlm.nih.gov/gtr/docs/submit/).
Submissions are integrated into the GTR database automatically. Novel data are flagged for curatorial review, and submitters are contacted to resolve any questions. The website is updated within 24 h of submission.
Submitters can also generate preliminary records for GTR either through migration of laboratory and test data from GeneTests or by completing a test submission template spreadsheet. The migration process allows submitters to take advantage of their data that is already maintained at NCBI through GeneTests (accessed through their GeneTests login ID and password) and to supplement the information using the GTR Submission User Interface before completing the registration in GTR. The spreadsheet mechanism is a semi-automated approach whereby a file containing all of the minimal and some recommended test fields is uploaded and the submission user interface is pre-populated with data for multiple tests. The submitter then reviews and submits each test individually. Submitters will also have the option to submit complete test and laboratory data as defined EXtensible Markup Language (XML) objects. The XML Schema Definition (XSD) to validate submissions is versioned, and the history of the versions and representative submission XMLs are provided from GTR’s FTP site (ftp://ftp.ncbi.nlm.nih.gov/pub/GTR/submission_templates/_README.html).
Quality of information submitted to GTR
A registry can expose areas where there is a lack of transparency but cannot independently evaluate data quality and accuracy. GTR states on the home page and throughout the website, ‘NIH does not independently verify information submitted to the GTR; it relies on submitters to provide information that is accurate and not misleading. NIH makes no endorsements of tests or laboratories listed in the GTR. GTR is not a substitute for medical advice. Patients and consumers with specific questions about a genetic test should contact a health care provider or a genetics professional.’ Similarly, as stated on its website, ‘GeneTests does not independently verify the accuracy of the service listings but relies on the listed laboratories and clinics to represent themselves accurately; users of GeneTests must independently verify the information listed herein.’
Submitters providing information to the GTR must agree to a Code of Conduct (http://www.ncbi.nlm.nih.gov/gtr/docs/code/) stating that they will uphold the integrity of the Registry through the submission of information that is accurate and not misleading; review and update the submitted information at least once a year and make no claims that NIH, HHS or the US government approves or endorses tests or other information submitted to the GTR. Users are encouraged to report any acts of inappropriate endorsement claims or any other breaches through a feedback form. A standard operating procedure is in place to resolve complaints, and the NIH reserves the right to remove the submitter’s records from the GTR.
GTR alerts users to data provided by submitters in three main ways. As explained in the section on GTR test pages, the bulk of the page (left and center) predominantly contains information submitted by test providers and the Discovery panel on the right contains information assembled by NCBI. Fields that are completed by submitters are marked as ‘Lab-provided’ in the adjacent tool tips. Finally, the help documentation and field definitions specify the sources of information in GTR.
Test quality is evaluated by a variety of proficiency testing programs throughout the world and GTR recommends reporting of such participation. The scope of this information includes proficiency testing providers and methods, and includes international providers such as the European Molecular Genetics Quality Network. Information about College of American Pathologists (CAP) proficiency testing is implemented as a set of nested choices on the submission portal. Data regarding New York State Clinical Laboratory Evaluation Program (CLEP) certification (license number, date and status) is elicited and users may filter on this feature. NCBI anticipates that over time, users will gauge tests based on the completion of these fields and providers will increasingly submit this information.
Curatorial review
GTR curators have training in medical genetics. They review new entries, manually curate practice guidelines and clinical resources and consult with content experts on matters such as defining phenotypes and the hierarchical relationship of terms. In general, GTR curators are cautious about representing any name as an alternate to another when conflicts exist among the sources used. Each novel name or term in a category that GTR maintains by standard lists (e.g. novel target-indication relationships, new variants and new gene names) is reviewed. Curators work closely with submitters and GTR users to ensure a smooth experience.
Database maintenance
The information in GTR is maintained both in a normalized database, and as XML specific to laboratories and tests. Data are processed for web retrieval daily by a process flow that includes identifying records that have been added or updated, and re-indexing them for retrieval. Automated daily jobs update automatically provided information from Gene, NCBI protein, dbSNP, GeneReviews and OMIM. Data from the HPO are updated weekly, and concepts from UMLS are updated with UMLS releases. The data from automated processes are integrated with submitted test data to maintain consistency on all tests.
Relationship of GTR to GeneTests
GeneTests is a pioneering NIH-funded resource developed at the University of Washington under the direction of Dr. Roberta Pagon and maintained at NCBI. GeneTests has multiple components including a Laboratory Directory and GeneReviews. The GeneTests Laboratory Directory is a compendium of laboratories that offer tests for genetic conditions; it does not provide a detailed description of specific tests nor report quality measures such as analytical or clinical validity. Laboratories with entries in GeneTests have permitted their data to be displayed in GTR and can migrate and supplement the information to complete GTR laboratory and test records. GTR and the GeneTests Laboratory Directory will overlap during a transition period anticipated to end in the first half of 2013. At that time, the GeneTests Laboratory Directory will be phased out.
GeneReviews will not be phased out with the GeneTests Laboratory Directory. Direct links from GeneReviews records to content in the GeneTests Laboratory Directory will be re-routed to GTR content in collaboration with the GeneTests staff. Condition- and gene-specific links to GTR will be enabled in sections of the right-hand column of individual GeneReviews. Searches of GeneReviews are now facilitated within GTR in a variety of ways as described below.
GTR includes information provided by laboratories to GeneTests in addition to fully registered laboratories and tests in GTR. All of the laboratory-submitted data shown in GeneTests has been shown within GTR since February 2012, labeled as ‘Displayed from GeneTests’. Differing views of test content during the transition period are described here: http://www.ncbi.nlm.nih.gov/gtr/docs/about/#currentcontent. The scope of the GTR extends beyond that of GeneTests by the explicit description of tests, the inclusion of pharmacogenetic tests and the characterization of certain analyses, such as HLA typing, as tests instead of services.
The transition from GeneTests to GTR without loss of information is enabled by automated mapping of data during interactive submission. Submitters may choose to complete only the minimal fields initially and return at a later time to submit recommended and optional fields. Recommended fields that are uncompleted are labeled ‘Not provided’ (e.g. clinical validity, clinical utility, target population, and proficiency testing). To the extent that users place value on the information contained in these fields, they may preferentially select tests for which these fields are completed and, over time, providers may choose to submit additional details. Thus it is anticipated that both the number of registered tests and the depth of information about individual tests will grow over time.
USER INTERFACE
Stakeholder input
NIH Director Francis S. Collins, M.D., Ph.D., publicly announced the launch of the GTR as part of Rare Disease Day on February 29, 2012. Before this, NIH had gathered broad input from stakeholders such as laboratory test developers, manufacturers and heath care providers. This was of prime importance in the development of the GTR, and continued stakeholder interaction is viewed as indispensable to optimizing the utility of the registry. Materials relevant to this process are maintained on the website of the NIH Office of Biotechnology Activities and are accessible through the GTR home page (http://oba.od.nih.gov/GTR/gtr_intro.html).
As part of the consultation process, NIH issued several requests for information and public comments, convened a public stakeholder meeting in November 2010 in conjunction with the annual meeting of the ASHG, and fulfilled the requirements by the Paperwork Reduction Act. Additional input was elicited at 19 meetings and teleconferences with stakeholders, 7 meetings of professional societies and from comments submitted through the GTR website.
Stakeholder input, in conjunction with guidance from the two GTR advisory groups, helped shape the GTR in several ways such as defining the scope of the registry, using a phased development approach, specifying the data elements and level of burden for providing these elements and determining the primary audience in the initial phase. Stakeholder input also helped NIH determine which data elements should be excluded from the initial phase of GTR such as test price, turn-around time and patent and licensing information.
Development
During the construction of the GTR, NCBI’s team of usability experts, web developers and content experts conducted usability testing with leaders from ACMG, AMP, ASHG, NSGC, the National Coalition for Health Professional Education in Genetics, genetic counseling training program directors, laboratory associations and laboratory personnel with expertise in biochemical, cytogenetic, molecular and pharmacogenetic testing. The GTR advisory groups provide guidance about display and usability on an ongoing basis.
General features of the website
GTR makes extensive use of hyperlinks to guide users to related information within GTR, other NCBI databases and external sites. Filters are provided on pages listing search results to help users find tests of interest. Tooltips accessed by a ‘?’ icon (Figure 1D) display the definition of specific fields and identify fields that are provided by submitters.
Test- and condition-specific pages feature a Discovery panel at the right (Figure 1B) that includes links of interest to the condition. The Reviews section at the top facilitates retrieval of context-specific records from the medical and scientific literature and features PubMed search tools tailored to genetic content. The Practice Guidelines section links to publications by professional organizations and to drug labeling information at the National Library of Medicine’s DailyMed website (http://dailymed.nlm.nih.gov/dailymed/). Concept-driven links to clinical, molecular and consumer resources are also provided. The juxtaposition of pertinent information enables GTR users to gauge the potential value of registered tests.
Home page and resource links
The GTR home page is the gateway to data, resources and information about GTR. Below the search bar, the left side of the page provides access to information about GTR (how to use the site, how to submit data, policy, structured feedback form, presentations schedule etc.). An extensive set of help documentation is available through the About GTR section, including how to search http://www.ncbi.nlm.nih.gov/gtr/docs/help/, filter, set preferences using MyNCBI, find services, find all registered laboratories and tests, submit data and access the GTR Fact sheet, presentations archive and YouTube instructional videos.
The right side enables searches of various resources outside GTR. Resources are organized by sections: Clinical Resources (GeneReviews, OMIM, Orphanet, NHGRI Talking Glossary), Locate a Genetics Professional, Consumer Resources (Genetics Home Reference, Office of Rare Diseases Research) and Molecular Resources (NCBI’s Genetics and Medicine resources). The criteria used to select links to non-government websites are described here: http://www.ncbi.nlm.nih.gov/gtr/docs/linkcriteria/.
Interactive data access
Data can be retrieved from the website either by searching all components of the database or focusing on one sector. The home page supports queries specific to Tests, Conditions/Phenotypes, Genes, Labs, GeneReviews and All GTR through tabs. Other pages provide all query types with the exception of GeneReviews through a menu at the right of the query box. Based on analyses of user behavior, the default query on the home page is set to Conditions/Phenotypes.
Queries typed by the user are aided by two tools: autocomplete and spell correction. As a query is typed, a list of terms in the database containing the typed characters is generated (autocomplete). If the desired query is displayed, a click on the term will result in the display of the record. The terms provided are customized to the type of query as summarized in Table 3. If a query contains a misspelled word that does not retrieve any record, a suggestion for the correctly spelled word is provided and the query processed for the new term.
Table 3.
Sector-specific search options
| Query type | Searchable by (autocomplete) |
|---|---|
| Tests | Test names, condition/phenotype names (with autocomplete), gene symbols and names (with autocomplete), protein names, laboratory names, directors, locations |
| Conditions/phenotypes | Condition/phenotype names (with autocomplete), protein names, analytes |
| Genes | Gene symbols and names (with autocomplete), condition/phenotype names (with autocomplete) |
| Labs | Laboratory names (with autocomplete), director and staff member names, locations, laboratory services, condition/phenotype names |
| All GTR | Condition/phenotype names (with autocomplete), gene symbols and names (with autocomplete), test names, protein names, analytes, laboratory names, director and staff member names, locations, laboratory services. The autocomplete category (condition or gene) is highlighted alongside the item name to differentiate similar terms and guide queries. |
| GeneReviews | Review title (with autocomplete), authors and terms contained within the PubMed excerpt |
Searching for GeneReviews
GeneReviews can be identified using GTR in several ways. In addition to the query tab mentioned above, users can use the link in the Clinical Resources section to go to the GeneReviews summary page in NCBI’s Bookshelf, where advanced searching is available. GeneReviews specific to a condition are also reported in the Discovery panel of the test page and condition/phenotype page. Additionally, GeneReviews are available through hyperlinks and colored chiclets on list result pages for conditions/phenotypes (see below) and the related diseases section of condition pages. GeneReviews searches are summarized here: http://www.ncbi.nlm.nih.gov/gtr/docs/help/#find_GeneReviews.
Search results
Query results display in the form of lists or the single result for the sector that was selected. Many paths are possible to retrieve the desired information; users are oriented by use of breadcrumbs summarizing their search path located beneath the search box (Figure 1A). An All GTR search result references all sectors. A special laboratory comparison page can be produced from a test list page by using the “Compare labs” function. The general layout and special features of these page types are summarized below.
Lists of results
An All GTR search produces a list organized into four sectors: Tests, Conditions/Phenotypes, Genes and Labs. The upper left side of the page provides the total number of data elements, which hyperlink to the analogous list pages; these counts are also listed within each section. Each section displays a maximum of three examples of each item in a standard layout similar to the corresponding list pages.
Ranking of search results is primarily determined by the search term chosen and the filtering strategy. Relevance sorting and the weighting of records that are more complete are described here: http://www.ncbi.nlm.nih.gov/gtr/docs/about/#ranking. Users may directly influence relevance by setting up preferred laboratories in MyNCBI, as explained in the section Limits, filters and preferences.
Each list produced for a specific sector of information displays the first 20 results of a query and summarizes the total number of results. The Test list also reports the number of tests for the number of conditions in the number of laboratories. All of the displayed data elements are provided by the submitting laboratory: Test name (hyperlinked to the test report page), Methods (chiclet and description), Analytical validity statement, Target population, Laboratory name (hyperlinked to the Laboratory page) and Directors.
The Conditions/Phenotypes list can be viewed as a set of descriptions (the default) or as related diseases. The description view is structured as a condition name (hyperlinked to the page specific to that condition), a brief description and a set of four hyperlinks to Tests, Genes, OMIM condition records and GeneReviews (links are active only when data are available). The related diseases view groups similar diseases together (each hyperlinked to the condition-specific page), provides a hierarchical representation and signals the availability of information by clicking on active colored chiclets for Clinical tests (C), Research tests (R), OMIM records (O) and GeneReviews (G).
Each item in the Gene list page displays the HGNC-approved gene symbol (hyperlinked to the gene item page) and gene name, followed by a gene description, list of alternate gene names, chromosome and location. A set of four hyperlinks (active when data are available) are provided to Tests, Diseases, OMIM gene and condition records, and ‘More about this gene’ links to the NCBI Gene record for the human gene.
Each item in the Laboratory list page is structured as the laboratory name (hyperlinked to the lab item page), institution name, list of directors, list of staff, number of tests, number of services and location. Hyperlinks are provided for the list of tests and the list of services.
Pages for specific tests, conditions, genes and laboratories
Test page
The test page (Figure 1) is the focal point of the GTR website. The two main sections of the page reflect the predominant sources of data for the page. The bulk of the page reports information submitted primarily by the test provider, organized under seven tabs. The Discovery panel at the right (Figure 1B) contains information assembled by NCBI. As is the norm in medicine and science, conflicting views may co-exist. This layout is intended to provide a comprehensive and balanced view of contextually relevant information from reputable sources along with resources for further exploration to facilitate an informed appraisal by the user.
Submitters determine the name of their test. The test fields and corresponding field requirements are enumerated in this table: http://www.ncbi.nlm.nih.gov/gtr/docs/fieldrequirements/#testrequirements. Data for test fields are distributed among the Overview, How To Order, Indication, Methodology, Performance Characteristics, Interpretation and Laboratory Contact tabs. Key features of the test pages include the unique GTR accession (Figure 1C), date the record was last updated, purpose of the test, methodologies used, targets of the test and a table summarizing what is tested, analytical validity, clinical validity, target population, clinical utility, proficiency testing, test codes, informed consent and genetic counseling requirements, protocol for interpreting variants of uncertain significance, use of the test sample for research purposes and sample reports. Submitted data are supplemented by NCBI (e.g. clinical features from sources such as OMIM and HPO). Links are provided to conditions and genes pertinent to the test and to the laboratory page.
The Test page content was determined following extensive input by GTR stakeholders and advisory groups, and may evolve as the testing landscape changes and additional feedback is provided. Elements such as turn-around time and cost were determined to be most readily kept up to date by participating laboratories on their own websites, and submitters may provide the direct URLs to this information (a recommended field) in the How to Order tab. Although Current Procedural Terminology (CPT) codes are not included at this time, GTR plans to include the new American Medical Association (AMA) Molecular Pathology (“MOPATH”) CPT codes, which provide more granularity than the stacking codes now in use.
Condition/phenotype page
This view orients the user to the specific condition for which tests may be offered by providing a description of disease characteristics. The Available Tests section contains a link to ‘See all available tests’, or the user can choose links to tests grouped by methodologies as a way to narrow the list. The Related Conditions section facilitates exploration of content through disease hierarchies and uses chiclets to signal the availability of content for clinical tests, research tests, OMIM records and GeneReviews. The Associated Genes section lists alternate gene symbols and gene names and links to the GTR gene page. The Clinical Features section provides additional details about the phenotypic features. Conditions for which limited content about disease characteristics is available can be further explored using the Related Conditions section and the Discovery panel.
Gene page
The gene-specific page is a subset of a report from NCBI Gene, customized for GTR. The complete Gene page is accessed by following the link ‘Go to complete Gene record for…’ at the upper right of the page. The GTR gene page provides a link to ‘See all available tests in GTR for this gene’. Condition-specific links to tests are available within the Associated Conditions section.
Lab page
This page organizes laboratory-specific information such as laboratory personnel, contact information, services, certifications (e.g. CLIA, CAP) licenses and links to the laboratory’s website.
All tests that the laboratory has registered in the GTR are provided as a scrollable menu in the Conditions and Tests section. A search by condition is provided for long test menus.
The Laboratory services field (http://www.ncbi.nlm.nih.gov/gtr/docs/submit/lab_info/#services) is a set of general services that may apply to several tests. Services important to everyday clinical operations such as insurance preauthorization, specialized methods such as preimplantation genetic diagnosis, broad analyses such as whole exome sequencing and offerings such as DNA banking may be selected by submitters. These choices display in the Lab page, are accessible through a hyperlink on Lab list pages and may be queried within the Lab or All GTR sectors.
Limits, filters and preferences
Given the breadth of data accessible from GTR, non-specific searches can return a large set of results. To enable a quick reduction of long result lists to specific records of interest, various filters and limits have been constructed for different sectors of information.
The list of conditions and phenotypes in GTR is extensive and the terms vary greatly with respect to the information available. Users may limit search results in a condition list by selecting checkboxes on the upper-left corner for the availability of GTR tests, GeneReviews or OMIM records. These three limits have a Boolean ‘and’ behavior—for example, checking the boxes for GTR Tests and GeneReviews delivers the subset of condition/phenotype results for which both types of records are available.
Searches that return more than one test result can be filtered in a number of ways using the filter section at the left of the page. The user may select one condition of interest and then click the Compare labs button to see laboratories that offer tests for that condition (see below). This function can be performed alone or in combination with additional filters including Test type (clinical or research), Test method (specific methodologies within the general categories Biochemical, Cytogenetic and Molecular), Test services, Lab certification (CLIA Certified, NY CLEP Certified and State Licensed) and Lab location (USA, specific states and other countries).
The Compare labs function supports review of laboratories offering tests for a specific condition. Methodologies offered by each laboratory are displayed horizontally by the use of colored ‘chiclets’ with a letter assigned for each method and a description beneath. Chiclets are vertically aligned for each laboratory on the list, allowing the user to scan the page for combinations of methods to plan their testing strategy. Services (e.g. Carrier testing, Prenatal testing) are also represented by chiclets. Laboratories can be sorted alphabetically or selected through checkboxes. Filters for Test method, Test services, Lab certification and Lab location are available in this view.
Searches that deliver a list of laboratories can be filtered for Lab certification, Test services and Lab location. Additionally, users can select preferred laboratories in GTR with their MyNCBI login, which implements preferential display of tests from selected laboratories at the highest rank in the test result page.
PROGRAMMATIC ACCESS
Data managed by GTR will be increasingly available by FTP with links provided from the home page. A README file (ftp://ftp.ncbi.nlm.nih.gov/pub/GTR/_README.html) is used to list content by category. The scope of the reports includes information about registered tests (data), standard terms, nomenclature for diseases, submission templates, documentation and presentations. GTR data are not currently accessible through NCBI’s E-utilities, although this is planned for 2013. Records will be rendered in XML.
Instructions for how to build links to GTR are provided here: http://www.ncbi.nlm.nih.gov/gtr/docs/linking/. URLs can be built based on the sector of information, and a set query can be used to interrogate content for all registered laboratories and tests. Websites currently referring to the GeneTests Laboratory Directory are encouraged to use this framework to route queries to the GTR before the end of the transition period.
Future phases for GTR
In the future, GTR will expand its scope to include tests for conditions with somatic mutations (e.g. malignancies), tests offered direct-to-consumer without a health care intermediary and tests that analyze and report results on whole exomes or whole genomes. Registration of manufacturers and test kits with description of non-proprietary information will be brought within the scope of GTR.
Research laboratories are described in GeneTests and this information is available in GTR. Submissions for research tests will soon be accepted to GTR. For the purposes of the GTR, a research test is defined as a test that is performed for the purpose of contributing to generalizable knowledge or for a laboratory to generate data to make technical improvements to a test.
Complete sequencing of an individual’s genome or exome is clinically available in several laboratories, though not yet a prevalent clinical activity. GTR currently facilitates the identification of laboratories offering these tests through a search of Laboratory services and will later enable registration of whole exome and/or whole genome tests. The description of tests using next-generation sequencing methods for gene panels is currently in scope.
Mechanisms to provide feedback to GTR
Extensive stakeholder input has shaped the GTR, and ongoing communication with the GTR staff is encouraged. A structured feedback form available on the home page elicits information about technical and content issues http://www.ncbi.nlm.nih.gov/gtr/feedback/. GTR provides an RSS feed (subscription provided from the home page) and a history of news releases (http://www.ncbi.nlm.nih.gov/projects/gtr/gtr_news.cgi).
Through ongoing participation by stakeholders, GTR aims to build a shared community resource which, over time can serve many purposes. Feedback from users about condition names and hierarchies will help refine the genetic lexicon. Submitter participation will enable a census of genetic tests, help assemble a compendium of practice guidelines relevant to genetics, build a comprehensive evidence base for genetic tests and enable health care providers to make informed appraisals of genetic tests.
FUNDING
Intramural Research Program of the NIH, National Library of Medicine and the NIH Office of the Director. Funding for open access charge: Intramural Research Program of the NIH, National Library of Medicine.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The GTR was developed by a team of database experts, programmers, web developers, usability experts and curators. Policy guidance was provided by the NIH Office of the Director. The public face of the GTR is the result of collaboration among multiple groups within the NCBI. We gratefully acknowledge the contributions of Baoshan Gu, Ricardo Villamarin-Salomon, George Riley, Melissa Landrum, and Kamen Todorov.
REFERENCES
- 1.Javitt G, Katsanis S, Scott J, Hudson K. Developing the blueprint for a genetic testing registry. Public Health Genomics. 2010;13:95–105. doi: 10.1159/000226593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zonno KD, Terry SF. Registry of genetic tests: a critical stepping stone to improving the genetic testing system. Genet. Test. Mol. Biomarkers. 2009;13:153–154. doi: 10.1089/gtmb.2009.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodenreider O, Mitchell JA, McCray AT. Evaluation of the UMLS as a terminology and knowledge resource for biomedical informatics. Proceedings/AMIA Annual Symposium. AMIA Symposium. 2002 pp. 61–65. [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's online mendelian inheritance in man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.UniProt, Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taschner PE, den Dunnen JT. Describing structural changes by extending HGVS sequence variation nomenclature. Hum. Mut. 2011;32:507–511. doi: 10.1002/humu.21427. [DOI] [PubMed] [Google Scholar]
- 12.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, Beahler C, Bird TD, Popovich B, Nesbitt C, et al. Genetests-geneclinics: genetic testing information for a growing audience. Hum. Mut. 2002;19:501–509. doi: 10.1002/humu.10069. [DOI] [PubMed] [Google Scholar]