Abstract
The identification of orthologous genes in an increasing number of fully sequenced genomes is a challenging issue in recent genome science. Here we present KEGG OC (http://www.genome.jp/tools/oc/), a novel database of ortholog clusters (OCs). The current version of KEGG OC contains 1 176 030 OCs, obtained by clustering 8 357 175 genes in 2112 complete genomes (153 eukaryotes, 1830 bacteria and 129 archaea). The OCs were constructed by applying the quasi-clique-based clustering method to all possible protein coding genes in all complete genomes, based on their amino acid sequence similarities. It is computationally efficient to calculate OCs, which enables to regularly update the contents. KEGG OC has the following two features: (i) It consists of all complete genomes of a wide variety of organisms from three domains of life, and the number of organisms is the largest among the existing databases; and (ii) It is compatible with the KEGG database by sharing the same sets of genes and identifiers, which leads to seamless integration of OCs with useful components in KEGG such as biological pathways, pathway modules, functional hierarchy, diseases and drugs. The KEGG OC resources are accessible via OC Viewer that provides an interactive visualization of OCs at different taxonomic levels.
INTRODUCTION
As the number of fully sequenced genomes is rapidly growing thanks to the advancement of next-generation sequencing technology, we face the necessity of analysing huge amount of genomic data in recent genome science. For example, 3402 organisms have been fully sequenced and 13 796 additional organisms are currently being sequenced according to the Genomes OnLine Database (GOLD) (1) as of writing this article. It is crucial to identify orthologous genes (orthologs) that are genes in different species and have branched from a single gene of their last common ancestor by speciation. The concept of orthologs plays a key role in functional annotation for newly sequenced genomes, because orthologs tend to have equivalent functions. In fact, functional annotation in many public databases is usually performed based on the sequence similarities of genes across different organisms. Those similar genes are often grouped together in a same ortholog cluster (OC) which naturally correlates with the functional classification. In practice, functional ontology classes such as Gene Ontology (GO) (2) are assigned to each gene. However, the reliability of the similarity-based functional annotation depends heavily on the similarity threshold and it should vary from gene family to family. OC delivers appropriate boundary to each sequence family by which the quality and scalability of functional annotation can be much improved.
From the viewpoint of systems biology, automatic pathway reconstruction is also of importance, because higher-level biological functions can be understood by pathways, or molecular interaction networks of gene products (e.g. metabolic pathways, regulatory pathways). KEGG PATHWAY is a typical pathway database and has a pathway-based assignment of orthologs named KEGG Orthology (KO), where each KO entry represents an ortholog group that is linked to a gene product in the KEGG pathway diagram (3). Once the KO identifiers (IDs) are assigned to genes in a genome, organism-specific pathways can be computationally generated, linking genomes to the biological systems. However, the KO entries are manually defined in KEGG, and a limited number of genes have been assigned to them. As the number of organisms stored into the KEGG database is exponentially growing in these days, manual assignment of the KO entries can be delayed. The use of automatically constructed OCs is expected to assist for the automatic pathway reconstruction in KEGG.
Computational identification of orthologs has been a longstanding problem in computational biology. The pioneering work is COG/KOG, which is based on the best-hit triangles between genes (4). COG/KOG has high-quality reference clusters, but it requires manual curation and lacks reproducibility. Considering a rapidly increasing number of fully sequenced genomes, it is necessary to automatically construct and update OCs. A serious problem of automatic OC construction is the difficulty of clustering a huge number of genes at once because of prohibitive computational cost. Recently, a variety of computational methods and databases have been developed to construct OCs from gene sequence similarity, and the previous methods can be categorized into multiple genome comparison or pairwise genome comparison. The multiple genome comparison approach is based on the clustering of genes across more than two organisms, similarly as COG/KOG. Examples include EGO/TOGA (5), MultiParanoid (6), OrthoMCL (7), OMAbrowser (8), MBGD (Microbial Genome Database) (9) and eggNOG (10), where the taxonomic information is also used in OMAbrowser and eggNOG. The pairwise genome comparison approach is based on the matching of genes between only two organisms. Examples include InParanoid (11) and Roundup (12), which are based on the bidirectional-best-hit and the reciprocal smallest distance, respectively. However, it is difficult to use the previously constructed OCs by the other groups in the KEGG database because of the data incompatibility problems and the insufficient coverage of organisms. There is, therefore, a strong incentive to develop methods to identify orthologous genes and to construct OCs every time complete genomes are newly sequenced.
In this article we present KEGG OC (KEGG Ortholog Cluster), a novel database of OCs based on the whole genome comparison. The OCs in KEGG OC were constructed by applying a novel clustering method to all possible protein coding genes in all complete genomes, based on their amino acid sequence similarities. The originality of our clustering algorithm lies in the use of a quasi-clique search (a variant of clique search) and the incorporation of phylogenetic information in the clustering. It is computationally efficient to calculate OCs, which makes it possible to regularly update the contents. KEGG OC has the following advantages over the existing databases in terms of organism coverage and compatibility with KEGG. (i) It consists of all fully sequenced genomes registered in KEGG, from a wide variety of organisms from three domains of life (eukaryotes, bacteria and archaea), and the number of organisms is the largest among the existing databases. (ii) It is compatible with KEGG by sharing the same set of genes and IDs, which leads to seamless integration of OCs with KEGG PATHWAY (biological pathways), KEGG MODULE (functional modules), KEGG BRITE (functional hierarchy), KEGG MEDICUS (diseases and drugs) and many more (3).
OVERVIEW OF KEGG OC
Construction of KEGG OC
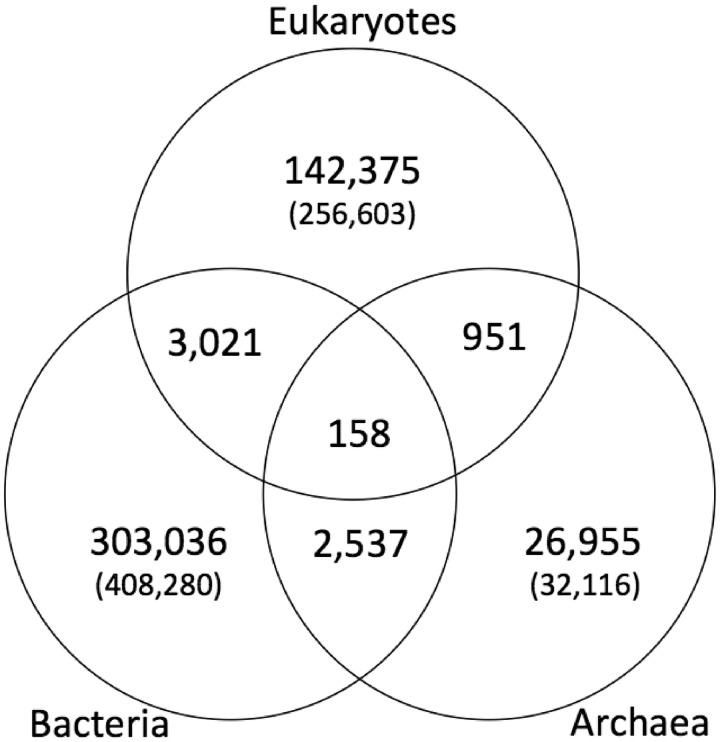
We applied the proposed method (see the ‘ALGORITHMS FOR CONSTRUCTING KEGG OC’ section) to cluster 8 357 175 genes in 2112 complete genomes (153 eukaryotes, 1830 bacteria and 129 archaea) stored in KEGG GENES of Release 62.0. KEGG OC contains the resulting 1 176 030 OCs, among which 696 999 are singletons (OCs consisting of a single gene), as of 9 July 2012. Figure 1 shows the distribution of OCs across three domains: eukaryotes, bacteria and archaea, where the number indicates the number of OCs consisting of multiple genes, whereas the number in parenthesis indicates the number of singletons. We compared the number of organisms and the number of genes in KEGG OC with those in other previous methods or existing databases: COG (13), KOG (13), EGO/TOGA (5), MultiParanoid (6), OrthoMCL (7), OMAbrowser (8), MBGD (9), eggNOG (10), InParanoid (11) and Roundup (12). Table 1 shows that the coverage of KEGG OC is much larger than those of other methods in terms of the number of organisms and the number of genes. We used 64 cores of a Xeon Processor (2.66 GHz) machine with 1024 cores and 16 TB memory. The whole calculation of the current version was finished within 2 weeks. The content of KEGG OC are regularly updated every 3 months.
Figure 1.
Distribution of OCs in KEGG OC across three domains: eukaryotes, bacteria and archaea. The number indicates the number of OCs consisting of multiple genes, whereas the number in parenthesis indicates the number of singletons (OCs consisting of a single gene).
Table 1.
Comparison of the numbers of fully sequenced organisms, eukaryotes, bacteria, archaea, genes and the last update date among COG, KOG, EGO/TOGA, MultiParanoid, OrthoMCL, OMAbrowser, MBGD, eggNOG, KEGG OC, InParanoid and Roundup
| Database/Method | The number of |
Last update year | ||||
|---|---|---|---|---|---|---|
| Organisms | Eukaryotes | Bacteria | Archaea | Genes | ||
| COG | 66 | 3 | 50 | 13 | 192 987 | 2003 |
| KOG | 7 | 7 | – | – | 60 759 | 2003 |
| EGO/TOGA | 88 | 88 | – | – | 618 733 | 2006 |
| MultiParanoid | 4 | 4 | – | – | 71 199 | 2011 |
| OrthoMCL | 150 | 98 | 36 | 16 | 1 398 546 | 2011 |
| OMAbrowser | 1211 | 124 | 994 | 93 | 5 701 696 | 2012 |
| MBGD | 1532 | 34 | 1382 | 116 | 5 415 388 | 2012 |
| eggNOG | 1133 | 121 | 943 | 69 | 5 214 234 | 2012 |
| KEGG OC | 2112 | 153 | 1830 | 129 | 8 357 175 | 2012 |
| InParanoid | 100 | 99 | 1 | – | 1 940 193 | 2009 |
| Roundup | 1786 | 226 | 1447 | 113 | 7 931 643 | 2011 |
OC Viewer for KEGG OC
The KEGG OC resources are accessible via OC Viewer at the KEGG OC website (http://www.genome.jp/tools/oc/). Users can input gene names, gene IDs (KEGG GENES IDs), KO IDs or annotation terms (e.g. thrA, eco:b0002, K12524 or dehydrogenase). Users can also search for OCs with an amino acid sequence of interest in OC Viewer, where BLAST is applied to searching for similar genes in KEGG GENES and then the hit genes are mapped to KEGG OC.
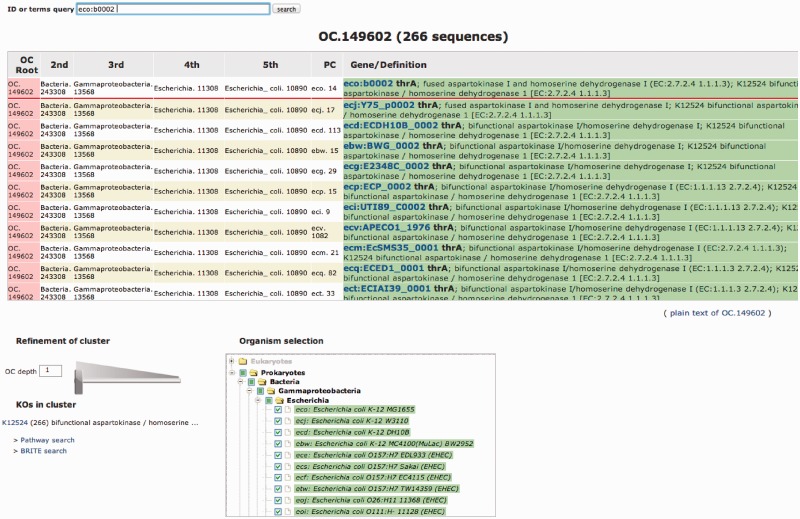
The outputs are the corresponding OCs, each of which provides the list of orthologous genes and their associated functional annotations in the cluster, and an interactive visualization at different taxonomic levels. Users can see, with this visualization, putative orthologous genes in taxon-based OCs [which we call taxon clusters (TCs)] in a hierarchical manner along a taxonomic classification and paralogous genes in paralog clusters (PCs) in each organism. Figure 2 shows an example of the output page of query ‘eco:b0002’ (an example of KEGG GENES ID for a gene of Escherichia coli K-12 MG1655) as an input. The ‘Pathway search’ and ‘BRITE search’ options enable the users to map KO IDs associated with the genes in the corresponding OCs onto pathway maps in KEGG PATHWAY and functional hierarchies in KEGG BRITE, respectively. The text file of the result can be downloaded in the tab-separated value (TSV) format.
Figure 2.
An example of the output page of OC Viewer of query ‘eco:b0002’ (an example of KEGG GENES ID for a gene of E. coli K-12 MG1655) as an input. The PC column shows the PCs (eco.14, ecj.17, ecd.113, etc.). These PCs are aggregated into a TC named Escherichia_col.10890 at the higher taxonomic level indicated in 5th column. As the aggregation of the TCs is iterated from the 5th column to the 2nd column in the OC table, these PCs are merged to the top-level cluster OC.149602. By using the slider at the bottom left, one can focus to arbitrary depth in the taxonomic tree indicated at the bottom right.
ALGORITHMS FOR CONSTRUCTING KEGG OC
Amino acid sequence similarity data
We obtained 8 357 175 genes from 2112 organisms (153 eukaryotes, 1830 bacteria and 129 archaea) from the KEGG GENES database, and evaluated the amino acid sequence similarity for 8 357 175× 8 357 175 gene pairs. We used the KEGG SSDB (Sequence Similarity DataBase: http://www.kegg.jp/kegg/ssdb/) as of 24 June 2012, which contains the information about amino acid sequence similarities among all protein coding genes in the complete genomes in KEGG. The similarities were computed by the SSEARCH program (14), and the Smith–Waterman (SW) similarity scores (15), symmetric similarity measures, were stored in SSDB. In this study we focus on gene pairs whose SW scores are ≥150.
An overview of our proposed method to construct OCs
We propose a new method to cluster all possible genes in all complete genomes based on the SW scores. The method consists of the following three steps:
Step 1. Construction of PCs. In each organism, we group genes that share higher similarity than any genes in the other organisms, borrowing the idea of the InParanoid algorithm (6). We then cluster all the genes in each organism by using the quasi-clique-based clustering (QCC) (The details of the QCC algorithm is described in the next subsection). The resulting clusters are referred to as PCs.
Step 2. Construction of TCs. In each taxon defined in the KEGG Organisms (http://www.kegg.jp/kegg/catalog/org_list.html), we group PCs that share higher similarity than any PCs in the other taxa (out groups). We then cluster all the PCs belonging to the same taxon by using QCC. The resulting clusters are referred to as TCs. We repeat the QCC-based clustering operation to cluster the newly generated TCs in a recursive way from lower taxa to higher taxa along a taxonomic tree until all taxa in each domain are merged.
Step 3. Construction of OCs. We group TCs that share enough similarity (with the minimal SW score ≥200) across three domains. We then cluster all the TCs by using QCC. The resulting clusters in the final stage are referred to as OCs.
In summary, the proposed method provides us with three types of gene clusters: (i) PCs at the level of each organism (e.g. human, chicken, fruit fly, E. coli K-12 MG1655, Methanocaldococcus jannaschii); (ii) TCs at the level of each taxon (e.g. mammals, birds, insects, Gammaproteobacteria, Euryarchaeota); and (iii) OCs at the level of all organisms from three domains (eukaryotes, bacteria and archaea). Note that each TC can be viewed by specifying the OC depth in OC Viewer.
QCC used in each step
We propose a heuristic clustering algorithm, which we name ‘quasi-clique-based clustering (QCC)’ after P-quasi complete graph (16). Note that QCC is used for clustering objects (e.g. genes, PCs, TCs) in each step. The QCC algorithm consists of the following five steps: First, we divide all objects into connected components in each of which objects are connected by paths that are constituted by object–object relations with the SW scores. Second, we represent each object by a neighbor profile in which each element corresponds to the minimal SW score of a gene pair that connects the object and the other object. Third, we evaluate the similarity between objects by computing the inner product of their neighbor profiles. Fourth, we apply a hierarchical clustering based on the computed neighbor profile similarities and obtain the dendrogram of objects. Finally, we divide the dendrogram based on the block enrichment along diagonal elements in the neighbor profile similarity matrix so that at least 50% of the object pairs are connected in each diagonal element.
More detailed explanation about the algorithm can be found in the Help page of OC Viewer (http://www.genome.jp/tools/oc/help.html).
DISCUSSION
A unique feature of KEGG OC is that it provides links from an OC to KO through the KEGG GENES entries of genes in each OC. The KO system is the basis for representing the KEGG reference pathway maps to be applicable to all organisms, and each KO entry represents a manually defined and context-dependent ortholog group. The automatically generated OCs tend to correspond to the manually curated KO entries at a reasonable level. For example, more than 90% of OCs including genes with the KO information corresponds to only one KO. The result suggests that OCs can be used as a basis of KO assignment to the KEGG GENES entries and as a complement resource of KO in order to explore anonymous protein families.
The KO assignment has been computationally and manually performed based on cross-species genome comparison using the KOALA (KEGG Orthology and Links Annotation) system (17). Recently, the information about OCs has been incorporated in the KOALA system to improve the process of KO assignment. A promising application of KEGG OC is to perform an automatic KO/GO assignment and pathway reconstruction for newly sequenced genomes, which will be made possible by using the KAAS (KEGG Automatic Annotation Server) system (18). An example of this application is shown as a function of CIPRO, the Ciona intestinalis protein database (19), where homologs were identified by KAAS with KEGG OC to help manual annotation by curators. These data are provided as links in CIPRO for users to obtain further biological implication.
Another example of the applications is shown in the ODB database (20), where genes comprising an operon are predicted by using orthologous information of KEGG OC. This property can also be used for functional annotation for OCs. The molecular functions of genes in each OC suggest possible annotations for the corresponding OC. If genes in an uncharacterized OC are clustered with genes in characterized OCs on a genome, the uncharacterized OC is estimated to have similar molecular functions with the characterized OCs.
The OCs can also be used for constructing phylogenetic profiles (21), which is useful for capturing the evolutionary expansion of protein functions/families. The OC-based phylogenetic profiles are used in the GENIES (GEne Network Inference Engine based on Supervised analysis) system (22), which predicts potential functional interactions between genes using a statistical learning method with gene–gene similarities computed from the OC-based phylogenetic profiles.
FUNDING
Japan Science and Technology Agency (in part). Funding for open access charge: Japan Science and Technology Agency.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Computational resources were provided by the Supercomputer system in the Bioinformatics Center, Institute for Chemical Research, Kyoto University. This article is dedicated to the memory of Masumi Itoh who passed away in July 2012.
REFERENCES
- 1.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Sultana R, Pertea G, Cho J, Karamycheva S, Tsai J, Parvizi B, Cheung F, Antonescu V, White J, et al. Cross-referencing eukaryotic genomes: TIGR Orthologous Gene Alignments (TOGA) Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexeyenko A, Tamas I, Liu G, Sonnhammer EL. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;22:e9–e15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- 7.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB. Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–D294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35:D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deluca TF, Cui J, Jung JY, St. Gabriel KC, Wall DP. Roundup 2.0: Enabling comparative genomics for over 1800 genomes. Bioinformatics. 2012;28:715–716. doi: 10.1093/bioinformatics/bts006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson WR. Empirical statistical estimates for sequence similarity searches. J. Mol. Biol. 1998;276:71–84. doi: 10.1006/jmbi.1997.1525. [DOI] [PubMed] [Google Scholar]
- 15.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda H, Ishihara T, Hashimoto A. Classifying molecular sequences using a linkage graph with their pairwise similarities. Theor. Comput. Sci. 1999;210:305–325. [Google Scholar]
- 17.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endo T, Ueno K, Yonezawa K, Mineta K, Hotta K, Satou Y, Yamada L, Ogasawara M, Takahashi H, Nakajima A, et al. CIPRO 2.5: Ciona intestinalis Protein Database, a unique integrated repository of large-scale omics data, bioinformatic analyses, and curated annotation, with user rating and reviewing functionality. Nucleic Acids Res. 2011;39:D807–D814. doi: 10.1093/nar/gkq1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuda S, Yoshizawa AC. ODB: a database for operon organizations, 2011 update. Nucleic Acids Res. 2011;39:D552–D555. doi: 10.1093/nar/gkq1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci., USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotera M, Yamanishi Y, Moriya Y, Kanehisa M, Goto S. GENIES: gene network inference engine based on supervised analysis. Nucleic Acids Res. 2012;40:W162–W167. doi: 10.1093/nar/gks459. [DOI] [PMC free article] [PubMed] [Google Scholar]