Abstract
MicroRNAs (miRNAs) are small RNAs ∼22 nt in length that are involved in the regulation of a variety of physiological and pathological processes. Advances in high-throughput small RNA sequencing (smRNA-seq), one of the next-generation sequencing applications, have reshaped the miRNA research landscape. In this study, we established an integrative database, the YM500 (http://ngs.ym.edu.tw/ym500/), containing analysis pipelines and analysis results for 609 human and mice smRNA-seq results, including public data from the Gene Expression Omnibus (GEO) and some private sources. YM500 collects analysis results for miRNA quantification, for isomiR identification (incl. RNA editing), for arm switching discovery, and, more importantly, for novel miRNA predictions. Wetlab validation on >100 miRNAs confirmed high correlation between miRNA profiling and RT-qPCR results (R = 0.84). This database allows researchers to search these four different types of analysis results via our interactive web interface. YM500 allows researchers to define the criteria of isomiRs, and also integrates the information of dbSNP to help researchers distinguish isomiRs from SNPs. A user-friendly interface is provided to integrate miRNA-related information and existing evidence from hundreds of sequencing datasets. The identified novel miRNAs and isomiRs hold the potential for both basic research and biotech applications.
INTRODUCTION
MicroRNAs (miRNAs) are a family of small RNAs, ∼22 nt in length, that act as key post-transcriptional regulators of gene expression, modulating the translational efficiency and/or the stability of target mRNAs. Small RNA sequencing (smRNA-seq), one of the next-generation sequencing (NGS) applications, enables detection and profiling of miRNAs with particularly high levels of sensitivity and accuracy (1). Furthermore, smRNA-seq allows discovery of previously uncharacterized miRNA species and has revealed unexpected complexity among miRNAs. These smRNA-seq discoveries include not only novel miRNAs but also a series of miRNA variants, termed isomiRs (2). In recent years, smRNA-seq datasets have grown rapidly and have been deposited in public databases such as the Gene Expression Omnibus (GEO) (3) and ArrayExpress (4). Exploration of these massive datasets, however, remains a daunting challenge, and an integrative meta-analysis of all smRNA-seq datasets has not yet been well performed.
For the detection and identification of novel miRNAs, smRNA-seq is very promising because it is not as time-consuming and expensive as small RNA cloning methods (5). Many sequencing software tools have been developed to identify novel miRNAs. Li et al. evaluated eight software tools, namely miRDeep (6), miRanalyzer (7), miRTRAP (8), MIReNa (9), mirTools (10), DSAP (11), miRNAkey (12) and mireap (13), based on their common features and key algorithms, and recommended the best tools in predicting novel miRNAs for different data types (5).
IsomiRs are commonly reported in deep sequencing studies (14–27) and are unlikely to be due simply to degradation or sequencing errors (25,28). These variants have been reported to be biologically relevant and functionally cooperative partners of canonical miRNAs (14,21). The variations present in IsomiRs can be grouped into three types: editing (nucleotide substitution), trimming and addition (29). The latter two types cause 5′ and 3′ end-length heterogeneity of miRNAs. Editing is a consequence of adenosine or cytidine deaminase activities and causes nucleotide changes at different positions of the mature miRNAs (15,16,28,30–33). It has previously been shown that several miRNAs, edited in the seed sequence and with an increased level of editing throughout development, result in diversifying target recognition (16). Trimming results in the shorter mature miRNAs compared with the canonical ones. The 3′-to-5′ exoribonuclease Nibbler has also been reported to control 3′ end processing of miRNAs in Drosophila (34,35). Non-template nucleotide additions at the 3′ end of miRNAs have been reported as the common form of miRNA enzymatic modification (18,21,28) and can influence miRNA stability (36) and the efficiency of target repression (37). It has further been revealed that the frequency of 3′ addition to specific miRNAs changes with differentiation of human embryonic stem cells (18). Several enzymes, such as MTPAP, PAPD4, PAPD5, ZCCHC6, ZCCHC11 and TUT1, have been reported to govern 3′ nucleotide addition to miRNAs (18,36–38).
The miR/miR* nomenclature has been used to represent the dominant and minor mature products of precursor miRNAs. However, several studies have reported that the arm that makes the dominant product can change in different tissues, stages and species (14,39–43). Such changes have been called ‘arm switching’ and are likely to be general (39). For example, Grimson et al. have reported an instance of developmental arm switching between the embryonic and adult stages of sponges for miR-2015 (43). Cloonan et al. also listed several miRNAs whose dominant arm changes over different human tissues (14). Therefore, in release 19 of the miRBase database (miRBase R19), the nomenclature for mature miRNAs now designates them as -5p and -3p, rather than miR/miR*, in all species.
In this study, we present the YM500 database, which includes pipelines for miRNA quantification, isomiR identification, arm switching discovery and novel miRNA prediction from smRNA-Seq data. YM500 contains the results of meta-analysis from hundreds of public smRNA-Seq datasets and dozens of in-house ones. YM500 aims to provide researchers with integrated miRNA-related information with various graphical visualization pages from hundreds of sequencing datasets via a user-friendly web interface.
MATERIALS AND METHODS
Data collection and pre-processing
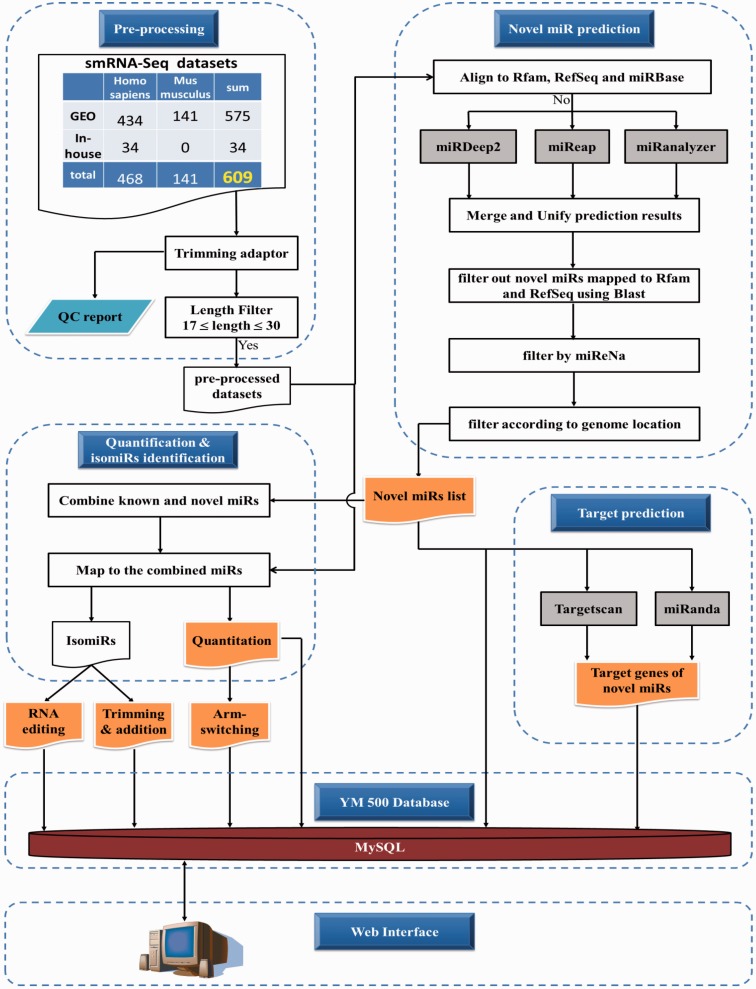
As shown under Pre-processing in Figure 1, there are 609 Illumina smRNA-Seq datasets, including 468 human and 141 mouse ones, in YM500. 34 out of 609 are in-house datasets, and the others are from the GEO public repository. All in-house data were deposited in the GEO database with an accession number of GSE39841. Detailed information for the datasets is provided in Supplementary Table S1. 3′ adaptors of the FASTQ raw data were trimmed, and the trimmed data were collapsed by the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit). A QC report, including length distribution, box plot of phred quality scores, etc., was then generated. For the public datasets containing only sequences and read counts, we transformed them into FASTA format and examined their length distribution. It has been suggested that if the smRNA-seq reads are abundant in 19∼24 nt, they are good for miRNA analysis (44). Thus, datasets which did not fit this criterion were discarded. For all datasets, reads shorter than 17 nt or longer than 30 nt were filtered out.
Figure 1.
Schematic representation of data processing.
Prediction of novel miRNAs and their downstream targets
It has been noted that there is no evidence that the miRNAs may represent fragments of mRNAs or other known RNA types (45). As shown in the Novel miRNA Prediction module in Figure 1, before predicting novel miRNAs, we used Bowtie (46) with options: -v 0 -f –norc to filter out reads that map to known miRNA precursors (miRBase) (45), other functional RNAs (Rfam) (47) and mRNAs (RefSeq) (48). Then, we adopted three prediction tools, namely miRDeep2, miReap and miRanalyzer, for predicting novel miRNAs. All of the prediction results were merged and unified to remove redundant records. According to our experience, there are still some predicted novel miRNAs that were mapped to known transcripts. We further applied BLAST to remove reads which were fully mapped to RefSeq or Rfam with identity >90%. To get a more reliable result, we also used miReNa as a second filter to filter out those that do not satisfy numerical Criteria I–V to describe a pre-miRNA in miReNa. About two-thirds of the putative miRNAs could not fulfil miReNa criteria and were filtered out. Finally, we filtered out the putative miRNAs located in exon regions (defined by RefSeq). These filtrations aimed to reduce the false positive rate. Filtration results are shown in Supplementary Figure S1. Most of the putative novel miRNAs that were predicted by multiple algorithms were preserved. There are 90 and 637 putative novel miRNAs predicted by three algorithms and any two algorithms, respectively (Supplementary Figure S1). Second structures of the stem-loop miRNA precursors were predicted by RNAfold (49). Furthermore, the target genes of these putative miRNAs were predicted by two well-known algorithms: TargetScan (50) and miRanDa (51). All information was stored in a local MySQL server.
miRNA quantification, isomiR identification and arm switching discovery
As shown in Figure 1, predicted novel miRNAs were combined with known miRNAs from miRBase R19. All pre-processed datasets were mapped to the combined miRNA list using Bowtie with options: -a -v 1 -S -f –norc, and then alignment results were produced in a BAM file format by SAMtools (52). The BAM files were processed by in-house JAVA software for miRNA quantification and isomiR identification. These analysis results were then stored in a local MySQL database. The arm switching events between two groups of samples were determined by two criteria: one was that the averaged RPKM (Reads Per Kilobase of transcript per Million mapped reads) value of a miRNA must be larger than 100. The second was that the ratios of two arms (5p/3p) in two groups must be significantly different according to the Student T-test with a P-value < 0.05 performed with the R language.
Validation of miRNA expression via stem-loop real-time PCR
Quantification of mature miRNAs was validated by a stem-loop real-time RT-qPCR system performed as previously described (53). Samples ranging from 100 ng to 1 μg of total RNA were used to perform reverse transcription (RT) using the RevertAidTM Reverse transcriptase kit (K1622; Fermentas, Glen Burnie, Maryland, USA) as directed by the manufacturer. Real-time PCR reactions were performed using MaximaTM SYBR Green qPCR Master Mix (K0222; Fermentas, Glen Burnie, Maryland, USA), and the specific products were detected and analysed using the StepOneTM sequence detector (Applied Biosystems, USA). Primers were designed on the basis of the sequenced miRNAs by using FastPCR (54). The miRNA expression data were normalized against U6 small nuclear RNA.
WEB INTERFACE
YM500 provides four interactive query interfaces (Expression, Novel miRNAs, isomiRs and arm-switching) and various graphical visualization pages to present the analysis results of hundreds of smRNA-Seq datasets.
Expression
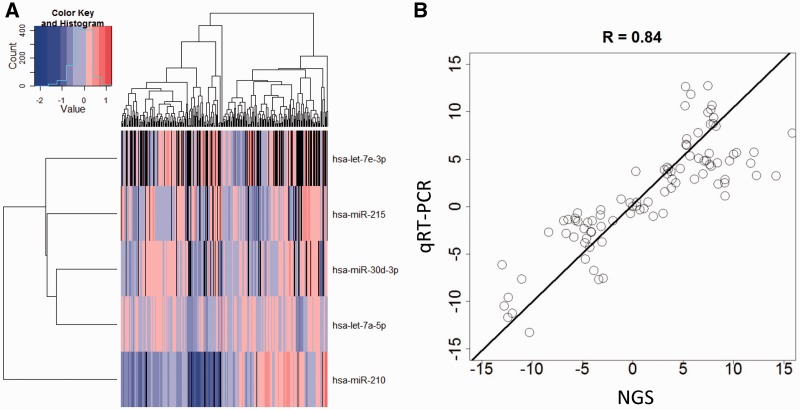
YM500 allows expression visualization according to a user’s customized selections. This feature helps users to select miRNAs according to ID lists or miRNA cluster definitions (Supplementary Figure S2A). For a query regarding a single miRNA, we provide the histogram expression in all samples and the expression profiles by tissue type of the specific miRNA. For a query regarding multiple miRNAs, samples can be selected according to the annotation (Supplementary Figure S2B). A recheck page (Supplementary Figure S2C) helps users to select the specific miRNA and samples for heatmap visualization (Figure 2A). A download link for the normalized expression data of selected samples and miRNAs is also provided for further analysis. Differential expression of 114 miRNAs was confirmed by RT-qPCR. The Pearson's correlation coefficient, R, between NGS analysis and RT-qPCR results was 0.84 (Figure 2B).
Figure 2.
miRNA expression. (A) The heatmap visualization for the expression of the selected miRNAs across samples. (B) The comparison of NGS and RT-qPCR in quantification of mature miRNAs.
Novel miRNAs
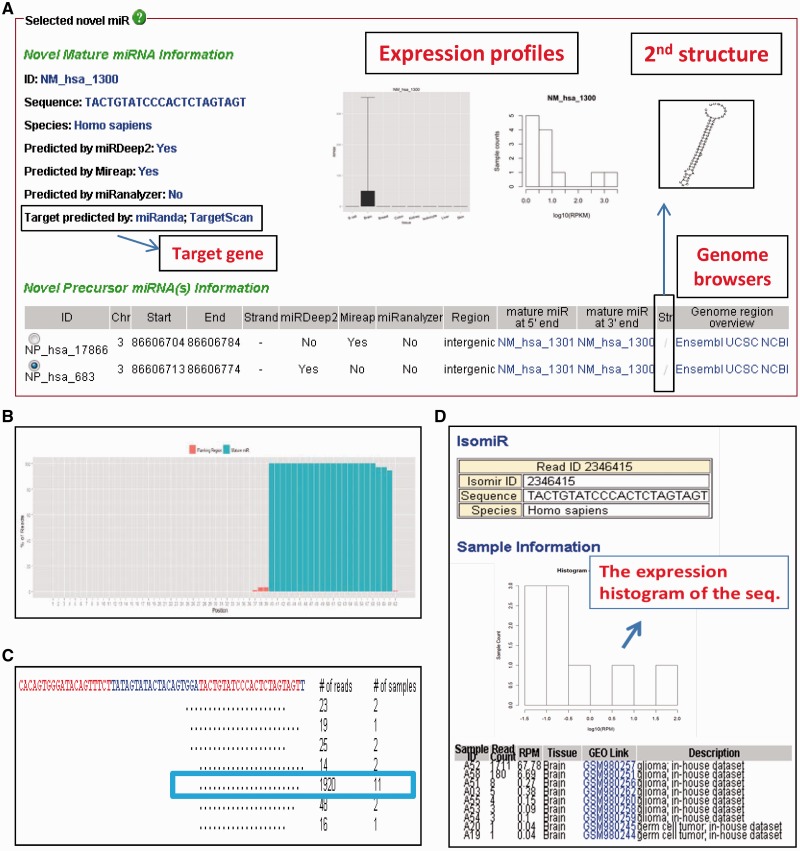
Whenever researchers identify potentially novel miRNAs, they can search for existing evidence of the novel miRNAs among the hundreds of samples in YM500. Novel miRNAs can be searched for according to the exact sequence of a mature miRNA or genomic location. Figure 3A illustrates the provided mature and precursor miRNA information, including the prediction algorithms, the predicted target genes, the expression profiles, the RNA secondary structures and the hyperlinks to three commonly used genome browsers. A density plot of reads that mapped to the stem-loop precursor indicates the percentage of reads overlapping the loci (Figure 3B). This provides an overview of length heterogeneity for a miRNA. Figure 3C shows a view of the deep sequencing reads and illustrates all sequences that map to the same novel miRNA. In the same figure, we also provide read numbers and numbers of datasets/samples in which a novel miRNA sequence was found. Reads can be filtered according to the number of mismatches to the hairpin sequence, the read count and the number of datasets/samples. Furthermore, each specific sequence (for example, the sequence in the rectangle in Figure 3C) has a hyperlink to a page (shown in Figure 3D) which contains the detailed information for the sequence, including the expression histogram, the raw counts and the RPM (Reads Per Million mapped reads) in each sample.
Figure 3.
Novel miRNA information. (A) The information for a novel mature miRNA (NM_hsa_1300; NM: novel mature miRNAs in YM500) and its precursor miRNA (NP_hsa_17866 and _683; NP: novel precursor miRNAs in YM500). (B) A density plot illustrating reads mapping to the putative precursor sequence (NP_hsa_683). Red and green bars indicate the flanking and the mature miRNA regions, respectively. (C) The view of deep sequencing reads. Each unique read is mapped to the putative precursor sequence (NP_hsa_683), with the putative mature sequence (NM_hsa_1300) highlighted in yellow. Dots indicate ‘perfect match.’ Numbers on the right show the read counts of each unique sequence and the number of samples in which this sequence was found. (D) The detailed information of the sequence labeled in the rectangle of (C). The expression histogram (middle) and the read counts and RPM of the sequence in the corresponding datasets (bottom) are shown.
IsomiRs
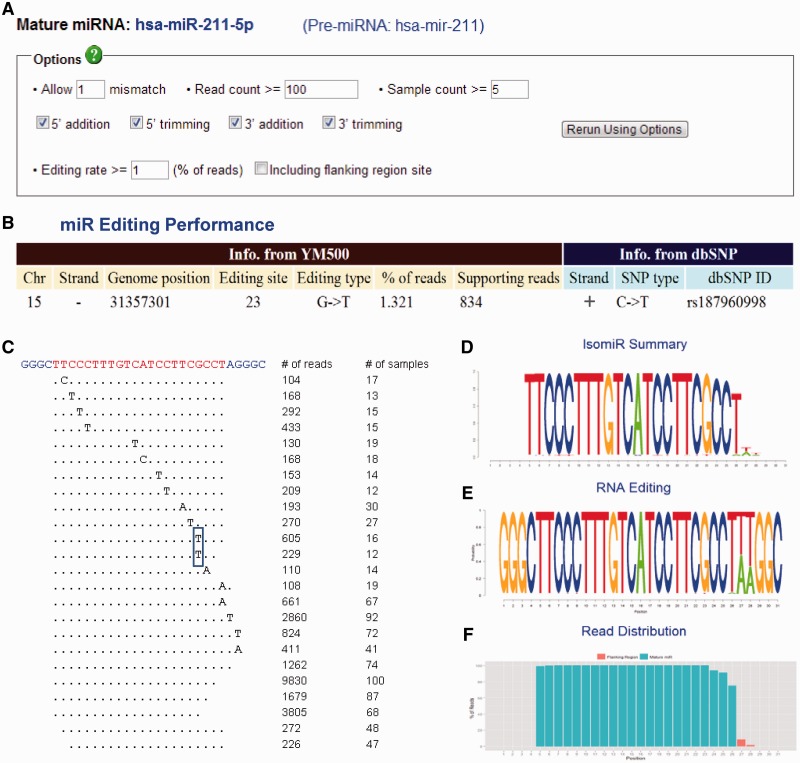
This section helps researchers to find the isomiRs of the known miRNAs in miRBase. As shown in Figure 4A, users can define the criteria of isomiRs according to number of mismatch, number of read counts, number of expressed samples and isomiR types (trimming or addition at 5′/3′ end). Figure 4B illustrates the information of edited sites, which are determined by the editing rate in Figure 4A. Editing information is also compared to dbSNP (Build 135). As shown in Figure 4B, both editing and an SNP were found at the 23rd base of hsa-miR-211-5p. However, the type of nucleotide alteration is different, indicating that such nucleotide substitution is a putative editing event rather than a common variant. All of the isomiRs defined in Figure 4A are detailed in Figure 4C, which indicates the mismatch sites, number of reads and number of samples containing the isomiR. Similar to the view of NGS data in the Novel miRNAs part (Figure 3C), each sequence in Figure 4C has a hyperlink to a page, shown in Figure 3D, which summarizes the details of the specific isomiR. Figure 4D shows the summary information of all isomiRs of a specific mature miRNA by a sequence logo format (where the height of each character is proportional to the total read counts of a miRNA). Figure 4E is similar to Figure 4D, but the height of each character is normalized to the read counts in each base and indicates the editing rate in each base. Similar to Figure 3B, Figure 4F is a density plot showing the distribution of reads in a mature miRNA and illustrates the length heterogeneity.
Figure 4.
IsomiR summarization. (A) A panel to define the criteria of isomiRs of a mature miRNA (e.g. hsa-miR-211-5p) from the precursor (hsa-mir-211). (B) The information of edited sites determined by the editing rate defined in (A). Editing information is also compared to that in dbSNP (Build 135). (C) The view of deep sequencing reads mapped to the sequence which contains the mature miRNA (hsa-miR-211-5p) and several flanking bases from the precursor (hsa-mir-211). Each unique isomiR is mapped to the sequence (top), with the mature sequence highlighted (yellow). Numbers on the right show the read count of each unique isomiR and the number of samples in which the isomiR was found. The rectangle indicates the putative editing site in (B). (D–E) The summary information for all isomiRs of a specific mature miRNA is illustrated in a sequence logo format. The number of the y-axis corresponds to the order of the sequencing on the top of (C). The height of each character is proportional to the total read counts (D) or to the read counts in each base (E) of the mature miRNA (hsa-miR-211-5p). (F) The density plot of reads mapping to the sequence on the top of (C). The red and green bars indicate the flanking and the mature miRNA regions, respectively.
Arm switching
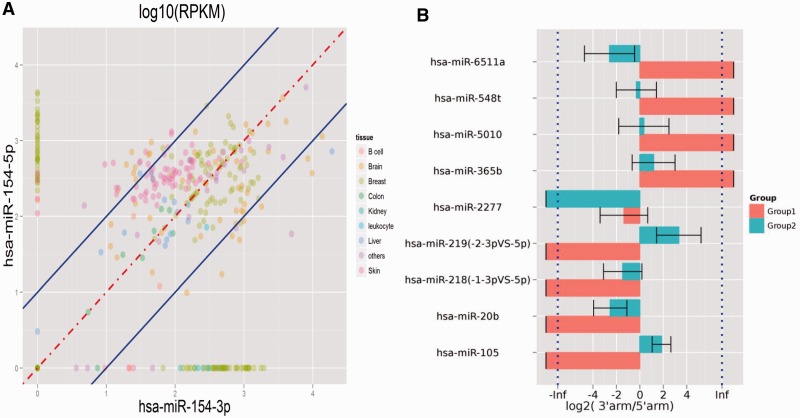
As far as arm switching is concerned, YM500 provides two ways to investigate this phenomenon. YM500 allows users to select a specific precursor miRNA and profiles the expression of two arms between samples and tissues (Figure 5A). This helps researcher to quickly view arm switching events in a specific miRNA species. Another method of illustration allows users to select two groups of samples, according to annotations for the database, and YM500 will identify precursor miRNAs whose dominant expression switches from one arm to the other between the two groups (Figure 5B).
Figure 5.
Arm switching. (A) The expression profiles of two arms of hsa-mir-154 between samples and tissues. (B) The precursor miRNAs with arm switching event, identified by YM500, between two groups of customer-defined samples.
DISCUSSION
YM500 integrates the analysis results of miRNA profiling/quantification, isomiR detection, novel miRNA prediction and arm switching identification. The reliability of in silico data were tested via experimental validation on in-house samples or by paper survey. For quantification, our miRNA profiling results were highly consistent to those of RT-qPCR, with a Person’s correlation coefficient 0.84 (Figure 2D). For novel miRNA discovery, we used multi-algorithms to explore as many putative miRNAs as possible and adopted several filtration steps to reduce false positive discoveries. The definition of ‘novel miRNA’ in YM500 is with respect to miRBase and we are collecting the information of ‘novel miRNA’ claimed by other references as another source of evidences but this task is undergoing. A dozen of miRNAs have been validated in our lab for their existence and expression patterns by RT-qPCR (Supplementary Table S2). Using ‘NM_hsa_1300’ as an example (Figure 3), wetlab evidence such as the RT-qPCR melting curve proved its existence (Supplementary Figure S3). However, RT-qPCR can prove the ‘existence’ of putative miRNAs. We use Ago1/2-mediated RNA-immunoprecipitation (RNA-IP) plus further sequencing as another line of evidence, and found that >1000 novel miRNAs in YM500 are indeed associated with the RNA-induced silencing complex (RISC) (unpublished data). For isomiRs, a U-to-G substitution in the ninth base of mmu-let-7a-5p (32) is discovered in YM500. Such substitution events (putative editing event) may result in a significant increase in stability of down-regulated targets (32). The second example is that we also discovered three reported A-to-I putative editing events in three miRNAs, which were the 7th, 8th and 9th bases of the mature product, and edited in 25%, 18% and 11% of the reads in (55), indicating that additional variability is tolerated in the functionally important seed region.
As for an example of arm switching, for hsa-mir-154 (Figure 5A), Cloonan et al. have reported that the expression of the two arms would be switched in different tissues (14). They demonstrated the switch between expression dominance from the 3p arm (ovary) to the 5p arm (brain and placenta). It has been shown that alternative mature miRNAs produced from the same precursor have different targeting properties and therefore different biological functions (56). Hence, the changes in arm choice of hsa-mir-154 might have significant functional consequences. The expression of a specific arm may dominate some tissue-specific functions. Besides, hsa-mir-144 has been reported as a cancer/disease marker in several studies (57–59) but our arm switching results show that it might have significant tissue-specificity. It may need to be further investigated the mechanisms of hsa-mir-144 which are related to cancer and tissue-specificity.
The advantage of using smRNA-Seq for miRNA researches is that smRNA-Seq can discover novel miRNAs and isomiRs. At this point, the number and functions of isomiRs remain unclear, but it has been reported that they might be quite prevalent in creatures. YM500 allows researchers to define the criteria of isomiRs, as well as providing existing evidence for various isomiRs from hundreds of smRNA-Seq datasets. A representative example of expression patterns and raw read counts in each smRNA-seq dataset is shown in Figure 4. We defined a candidate isomiR by the criteria shown in Figure 4A (allow one mismatch; the read count and sample count are at least 100 and 5, respectively). At the same time, the editing event is defined by the editing rate at least 1% of the total reads (pass the criteria) mapped to hsa-miR-211-5p. (Please note that all of criteria could also be customer-defined). In this case, 13 sites of the canonical miRNAs have isomiRs with mismatch, but there is only one putative editing event in the 23rd base with 834 supporting reads in more than a dozen of datasets (Figure 4B and C). Besides, there are two distinct isomiRs that have the same editing site (the rectangle in Figure 4C). We also provide the detail information of each isomiR via hyperlinks on web interface. These isomiRs still need extensive wetlab validations to prove their existence and functionality. Especially, the editing events need genomic evidences from the same sample to rule out novel SNPs or mutations. However, YM500 is a screening tool to help researchers reduce the numbers of candidate isomiRs and could be severed as a line of evidences for their existence. We believe these customer-defined criteria and selections would help researchers to exclude most sequencing errors and other artifacts.
The same holds true for novel miRNAs. When a small RNA is consistently detected in various datasets, it does constitute autonomous evidence that it is prevalent and thus the likely result of a specific biogenesis (6). As shown in Figure 3, for a putative novel miRNA, YM500 provides the expression profiles, the prediction algorithms, the sequences, the counts mapping to the miRNA, the secondary structure, etc. This information helps researchers to evaluate the prediction result. For example, according to the suggestions of Kozomara and Griffiths-Jones (45), the pattern of reads focusing on the mature region (Figure 3B) supports a high-confidence miRNA annotation with multiple reads (10–20 as cutoffs) from independent datasets. In contrast, the pattern of reads overlapping the sequence of a putative miRNA does not support the annotation of a miRNA, with multiple offset reads distributed across the locus (45). Besides, most reads mapping to a given mature miRNA annotation should have the same 5′ end whereas the 3′ end may be significantly more variable (45). YM500 helps researchers to check these characteristics via the presentation page (Figure 3). For isomiRs and novel miRNAs, YM500 provides existing evidence from hundreds of smRNA-Seq datasets. Whenever researchers find interesting results (such as some specific isomiRs and novel miRNAs) in their own datasets, they can validate their results in YM500.
In comparison to other databases related to smRNA-seq, including miRBase, deepBase (60) and the isomiRs database of Lee et al. (24), YM500 analyse miRNA-related information from many dimensions, and datasets included in meta-analysis are more abundant. The deepBase database contains 185 smRNA-Seq datasets for seven organisms and is also a platform for annotating and discovering small and long non-coding RNAs (miRNAs, siRNAs, piRNAs, etc.) from next-generation sequencing data. The isomiRs database of Lee et al. contains only 18 smRNA-Seq datasets for two organisms and lists only the isomiR information in a single sample per query. Although miRBase covers much more species, miRBase does not include expression profiles of known miRNAs, RNA editing, arm switching or miRNA prediction results. For novel miRNA, YM500 adopts four different algorithms and provides target prediction, expression profiles of novel miRNA and hyperlinks to genome browsers. deepBase lists only the results of novel miRNA predicted by miRDeep, and no other additional information is provided. Besides, there are only 79, 9 and 5 human smRNA-seq datasets in miRBase, deepBase and the isomiRs database of Lee et al., respectively. YM500 contains 468 human datasets (from public databases or in-house), which is the most comprehensive collection so far. Another unique part of YM500 is that our interactive web interface and customer-defined criteria also help researchers to retrieve these four types of analysis results. Finally, to our best knowledge, there is no other resource providing arm switching information. Comparing with these databases, YM500 provides a flexible web interface, more enhanced resolution and novel findings owing to the integrated pipelines for miRNA (including miRNA expression, IsomiRs, novel miRNAs and arm-switching) and the large number of smRNA-Seq datasets of various tissue/cell types.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–3.
FUNDING
Funding for open access charge: National Science Council [NSC98-2320-B-010-020-MY3, NSC99-2320-B-350-003-MY3, NSC100-2627-B-010-007, NSC100-2314-B-075-066-MY2 and NSC101-2320-B-010-059-MY3]; Taipei Veterans General Hospital [V101E2-008 and Cancer Excellence Center Plan, DOH101-TD-C-111-007]; National Research Program for Biopharmaceuticals [DOH101-TD-PB-111-TM007]; National Yang-Ming University via the Ministry of Education, Aim for the Top University Plan; UST-UCSD International Center for Excellence in Advanced Bioengineering sponsored by the Taiwan NSC I-RiCE Program [NSC100-2911-I-009-101, in part].
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhou L, Li X, Liu Q, Zhao F, Wu J. Small RNA transcriptome investigation based on next-generation sequencing technology. J. Genet. Genomics. 2011;38:505–513. doi: 10.1016/j.jgg.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q, Shen B. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4298–4305. doi: 10.1093/nar/gks043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011;39:W132–W138. doi: 10.1093/nar/gkr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrix D, Levine M, Shi W. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol. 2010;11:R39. doi: 10.1186/gb-2010-11-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathelier A, Carbone A. MIReNA: finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics. 2010;26:2226–2234. doi: 10.1093/bioinformatics/btq329. [DOI] [PubMed] [Google Scholar]
- 10.Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun Z, Wu J. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38:W392–W397. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang PJ, Liu YC, Lee CC, Lin WC, Gan RR, Lyu PC, Tang P. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res. 2010;38:W385–W391. doi: 10.1093/nar/gkq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronen R, Gan I, Modai S, Sukacheov A, Dror G, Halperin E, Shomron N. miRNAkey: a software for microRNA deep sequencing analysis. Bioinformatics. 2010;26:2615–2616. doi: 10.1093/bioinformatics/btq493. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Li Q, Wang J, Guo X, Jiang X, Ren Z, Weng C, Sun G, Wang X, Liu Y, et al. Identification and characterization of novel amphioxus microRNAs by Solexa sequencing. Genome Biol. 2009;10:R78. doi: 10.1186/gb-2009-10-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloonan N, Wani S, Xu Q, Gu J, Lea K, Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danecek P, Nellaker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, Flint J, Durbin R, Keane TM, Adams DJ. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012;13:r26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekdahl Y, Farahani HS, Behm M, Lagergren J, Ohman M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 2012;22:1477–1487. doi: 10.1101/gr.131912.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vesely C, Tauber S, Sedlazeck FJ, von Haeseler A, Jantsch MF. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 2012;22:1468–1476. doi: 10.1101/gr.133025.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen B, Zhang B, Luo H, Yuan J, Skogerbo G, Chen R. Distinct microRNA subcellular size and expression patterns in human cancer cells. Int. J. Cell Biol. 2012;2012:672462. doi: 10.1155/2012/672462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guduric-Fuchs J, O'Connor A, Cullen A, Harwood L, Medina RJ, O'Neill CL, Stitt AW, Curtis TM, Simpson DA. Deep sequencing reveals predominant expression of miR-21 amongst the small non-coding RNAs in retinal microvascular endothelial cells. J. Cell. Biochem. 2012;113:2098–2111. doi: 10.1002/jcb.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo L, Li H, Liang T, Lu J, Yang Q, Ge Q, Lu Z. Consistent isomiR expression patterns and 3′ addition events in miRNA gene clusters and families implicate functional and evolutionary relationships. Mol. Biol. Rep. 2012;39:6699–6706. doi: 10.1007/s11033-012-1493-3. [DOI] [PubMed] [Google Scholar]
- 22.Burroughs AM, Kawano M, Ando Y, Daub CO, Hayashizaki Y. pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5′/3′ arm variation including concomitant cleavage and polyuridylation patterns. Nucleic Acids Res. 2012;40:1424–1437. doi: 10.1093/nar/gkr903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H, Arcila ML, Li Z, Lee EJ, Henzler C, Liu J, Rana TM, Kosik KS. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012;40:5864–5875. doi: 10.1093/nar/gks247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA. 2011;17:1795–1803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voellenkle C, van Rooij J, Guffanti A, Brini E, Fasanaro P, Isaia E, Croft L, David M, Capogrossi MC, Moles A, et al. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA. 2012;18:472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humphreys DT, Hynes CJ, Patel HR, Wei GH, Cannon L, Fatkin D, Suter CM, Clancy JL, Preiss T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012;7:e30933. doi: 10.1371/journal.pone.0030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pantano L, Estivill X, Marti E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38:e34. doi: 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reid JG, Nagaraja AK, Lynn FC, Drabek RB, Muzny DM, Shaw CA, Weiss MK, Naghavi AO, Khan M, Zhu H, et al. Mouse let-7 miRNA populations exhibit RNA editing that is constrained in the 5′-seed/cleavage/anchor regions and stabilize predicted mmu-let-7a:mRNA duplexes. Genome Res. 2008;18:1571–1581. doi: 10.1101/gr.078246.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alon S, Mor E, Vigneault F, Church GM, Locatelli F, Galeano F, Gallo A, Shomron N, Eisenberg E. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012;22:1533–1540. doi: 10.1101/gr.131573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr. Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr. Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S, Hui JH, Marco A, Ronshaugen M. MicroRNA evolution by arm switching. EMBO Rep. 2011;12:172–177. doi: 10.1038/embor.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marco A, Hui JH, Ronshaugen M, Griffiths-Jones S. Functional shifts in insect microRNA evolution. Genome Biol. Evol. 2010;2:686–696. doi: 10.1093/gbe/evq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li SC, Liao YL, Ho MR, Tsai KW, Lai CH, Lin WC. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics. 2012;13(Suppl. 1):S13. doi: 10.1186/1471-2164-13-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li SC, Liao YL, Chan WC, Ho MR, Tsai KW, Hu LY, Lai CH, Hsu CN, Lin WC. Interrogation of rabbit miRNAs and their isomiRs. Genomics. 2011;98:453–459. doi: 10.1016/j.ygeno.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinform. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 51.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalendar R, Lee D, Schulman AH. Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Genomics. 2011;98:137–144. doi: 10.1016/j.ygeno.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Parts L, Hedman AK, Keildson S, Knights AJ, Abreu-Goodger C, van de Bunt M, Guerra-Assuncao JA, Bartonicek N, van Dongen S, Magi R, et al. Extent, causes, and consequences of small RNA expression variation in human adipose tissue. PLoS Genet. 2012;8:e1002704. doi: 10.1371/journal.pgen.1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marco A, Macpherson JI, Ronshaugen M, Griffiths-Jones S. MicroRNAs from the same precursor have different targeting properties. Silence. 2012;3:8. doi: 10.1186/1758-907X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akiyoshi S, Fukagawa T, Ueo H, Ishibashi M, Takahashi Y, Fabbri M, Sasako M, Maehara Y, Mimori K, Mori M. Clinical significance of miR-144-ZFX axis in disseminated tumour cells in bone marrow in gastric cancer cases. Br. J. Cancer. 2012;107:1345–1353. doi: 10.1038/bjc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaedcke J, Grade M, Camps J, Sokilde R, Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi BM, Moller S, et al. The rectal cancer microRNAome - microRNA expression in rectal cancer and matched normal mucosa. Clin. Cancer Res. 2012;18:4919–4930. doi: 10.1158/1078-0432.CCR-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu H, Li H, Zhang L, Luan H, Huang T, Wang L, Fan Y, Zhang Y, Liu X, Wang W, et al. Diagnostic role of microRNA expression profile in the serum of pregnant women with fetuses with neural tube defects. J. Neurochem. 2012;122:641–649. doi: 10.1111/j.1471-4159.2012.07812.x. [DOI] [PubMed] [Google Scholar]
- 60.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]