Abstract
TFClass (http://tfclass.bioinf.med.uni-goettingen.de/) provides a comprehensive classification of human transcription factors based on their DNA-binding domains. Transcription factors constitute a large functional family of proteins directly regulating the activity of genes. Most of them are sequence-specific DNA-binding proteins, thus reading out the information encoded in cis-regulatory DNA elements of promoters, enhancers and other regulatory regions of a genome. TFClass is a database that classifies human transcription factors by a six-level classification schema, four of which are abstractions according to different criteria, while the fifth level represents TF genes and the sixth individual gene products. Altogether, nine superclasses have been identified, comprising 40 classes and 111 families. Counted by genes, 1558 human TFs have been classified so far or >2900 different TFs when including their isoforms generated by alternative splicing or protein processing events. With this classification, we hope to provide a basis for deciphering protein–DNA recognition codes; moreover, it can be used for constructing expanded transcriptional networks by inferring additional TF-target gene relations.
INTRODUCTION
Most eukaryotic transcription factors (TFs) regulate transcription through binding to defined cis-regulatory elements in promoters, enhancers, silencers and other regulatory regions. These regions seem to be subject to a structurally as well as a temporally hierarchical organization. Some of their constituents, when co-occurring in a proper distance and orientation, may form composite modules as one important intermediate level between individual cis-regulatory elements and the whole region. The TFs binding to these modules may be additionally engaged in protein–protein interactions among each other, resulting in synergistic effects (1), as was systematically collected in the TRANSCompel database (2).
Among the effects of their binding may be to foster the formation of the basal transcription complex through contacts to general TFs (1), or to trigger chromatin remodeling through DNA or histone modifications; as for the latter, however, it may be arguable in which case the TF binding is the cause or the consequence of histone methylation or acetylation events (3). Concepts about pioneer factors (3), master TFs (4) or seed-site binding factors (5) have been developed and may provide a clue to understand how the assembly of proteins and their dynamics operate.
To exert their function to activate or repress transcription of gene(s), TFs have to recognize the place in the genome where they should bind to. For this, they are equipped with DNA-binding domains (DBDs), the characteristics of which determine the usually relaxed DNA-binding specificity of eukaryotic TFs. Since reading out the regulatory instructions encoded in the genomic nucleotide sequence is the key process for activating genetic programs in a highly controlled and subtly tuned manner, the problem of deciphering the protein–DNA recognition code has been tackled and solved to some extent for a few groups of DBDs, such as zinc finger proteins, nuclear receptors or helix-turn-helix factors (6). One aim of such attempts is to predict the DNA-binding specificity of yet uncharacterized proteins (7), which could largely benefit from a comprehensive classification of TFs according to the structural features of their DBDs.
To this end, Harrison in 1991 (8) has published a first and comprehensive taxonomy of DBDs, which comprised four groups (helix-turn-helix, zinc-binding, basic leucine zipper (bZIP) and beta-ribbon domains). Most of the factors and many of the principal topologies were not yet known at that time. A specific ‘census of human TFs’ was published by Vazquerizas et al. (9). Here, human TFs were assigned to 23 TF families, which were defined by considering parent–child relationships from the InterPro database (10). The principles of this classification were already published in 2000 (11). In the recently published AnimalTFDB, the authors made a comprehensive attempt to classify TFs according to their DBDs (12). They identified 71 animal TF families, which were then assigned to the early TRANSFAC superclasses. TFCat aims to provide a comprehensive catalog of human and mouse TFs, classified into a taxonomy according to their basic function, and in altogether seven defined (plus two undefined) groups with 39 families (13).
In 1988, we have started to collect information about individual, experimentally validated TF-binding sites (14), an attempt which then resulted in the TRANSFAC database (15,16). Soon, the individual binding sites were used as training sets to construct a library of positional weight matrices (PWMs), which were used by several tools to predict potential transcription factor binding site in DNA sequences, such as MatInspector or MATCH (17,18). In parallel, a TF classification scheme was developed in 1997 that was based on the properties of the DBDs of the TFs known at that time (19). The classification itself was later refined (20). Since then, a large number of TFs were discovered, for instance as a result of the international genome sequencing projects. Of even greater impact for the structure of the classification, a number of principally new DBD structures were discovered. In this contribution, an attempt is made to investigate whether the previously published classification scheme still applies and to propose a correspondingly revised TF classification. According to our experiences, when doing such a classification across multiple species, a certain bias is easily introduced for those TF groups that have been more intensely studied. More seriously, the distances between orthologous proteins of different biological species, which obviously increase with the evolutionary distance, and those between paralogous TFs, which may also reflect functional divergence, will be mixed up and may spoil the conclusions about functional (dis)similarity of the TFs encoded by one genome. To avoid such problems with sorting out paralogous and orthologous relationships, the classification will be done here for human TFs only.
MATERIALS AND METHODS
Data sources
Domain assignments and protein sequences were taken from UniProt (21). Information about isoforms was collected from UniProt, last update done using release 2012_07 (21), and TRANSFAC, with the last update using release 2012.2 (16). 3D structures were obtained from the PDB database (22), generally used as entry point to retrieve the original publications.
Domain annotation
All DBD annotations taken from UniProt were manually validated and, when necessary, corrected. After multiple sequence alignments (see next paragraph), the domain borders were N- and C-terminally trimmed to the consensus borders.
Sequence comparisons
The retrieved DBDs and/or the full-length protein sequences were subject to multiple alignments, usually by Clustal Omega (23), and subsequent cluster analyses as implemented at UniProt/Swiss Institute of Bioinformatics (SIB), or ClustalW (24) at the European Institute of Bioinformatics (EBI) or in the MEGA4 software package (25). In many cases, different clustering algorithms [neighbor joining, maximum parsimony, minimum evolution or the unweighted pair group method with arithmetic mean (UPGMA)] were applied for tree constructions and compared. They were routinely compared with the results obtained with the SATCHMO program (26). We applied these different algorithms in order to exploit different aspects of sequence similarities, with different underlying assumptions. They refer more or less explicitly to phylogenetic relation, and have been mostly applied to reveal such relations. Although being derived from a common ancestor is not a necessary prerequisite for classifying objects according to their similarity, we feel that it may be a reasonable hypothesis to assume that there are phylogenetic relations between the TFs, at least those of one (super-) class.
Web interface
The classification data were transferred into an Open Biomedical Ontologies (OBO) format. The classification is then visualized dynamically as a tree in a web application based on JavaServer Faces (JSF). The data for browsing and the search are retrieved from our OBA (‘ontology based answers’) server in JavaScript Object Notation (JSON) format (27).
STRUCTURE OF THE CLASSIFICATION
Rank definitions
The general idea of the classification scheme is to provide a hierarchical system of taxa, inspired by both the taxonomy of biological species on the one and the enzyme catalog (28) on the other side. In accordance with the latter, a four-level taxonomy was proposed (19), comprising the ranks superclass, class, family and subfamily, the latter as an optional category (Table 1).
Table 1.
Rank definitions
| Level | Rank denomination | Definition | Example |
|---|---|---|---|
| 1 | Superclass | General topology of the DBD | Zinc-coordinating DBDs (Superclass 2) |
| 2 | Class | Structural blueprint of the DBD | Nuclear receptors with C4 zinc fingers (Class 2.1) |
| 3 | Family | Sequence and functional similarities | Thyroid hormone receptor-related factors (NR1) (Family 2.1.2) |
| 4 | Subfamily | Sequence-based subgroupings | Retinoic acid receptors (NR1B) (Subfamily 2.1.2.1) |
| 5 | Genus | TF gene | RAR-α (Genus 2.1.2.1.1) |
| 6 | Factor ‘species’ | TF polypeptide | RAR-α1 (Species 2.1.2.1.1.1) |
TF superclasses are defined according to the general topology of their DBDs and the mode of their interaction with the target DNA sequence. This definition may be expanded to an adjacent di-(or multi-)merization domain if the resulting protein–protein interactions are a prerequisite for the DNA binding or influence the DNA-binding specificity.
The class level was the primary one that was defined very early in the TRANSFAC database (29,30). At this level, structural and sequence similarities together constitute larger groups of TFs that share a structural DNA-binding motif which can be traced back to sequence similarities.
Each class comprises one or several families, which are primarily defined on the basis of sequence similarities of their DBDs, again following the idea that similar DBDs may interact with related DNA sequences. To identify these family relations, a number of multiple sequence alignment methods were employed (see ‘Materials and Methods’ section).
In general, family definitions were accepted as sufficiently robust only if they were consistently suggested by all (or most) algorithms and implementations applied. Optionally, some families have been subdivided into subfamilies, if there were clear subgroupings recognizable during the cluster analysis. It should be noticed that the exact criteria to assign individual (groups of) factors to a family or subfamily vary among the different classes, since they have to reflect the particularities of the specific class they belong to.
Beyond the optional subfamily level, two more levels were defined, which represent physically definable entities (i.e. genes and gene products) and, being inspired by the taxonomical systems of biological species, have been termed ‘genera’ and ‘species’ (or ‘molecular species’, to avoid confusion with biological species). Thus, all TFs encoded by one gene have been put into taxa on the fifth level of the TF classification, whereas the different products of one gene represent the sixth level, i.e. that of molecular species, only if there are several isoforms described.
Naming conventions
As for naming of the classes, families and subfamilies, the following rules have been established.
Families are mostly named ‘x-related factors’, according to one of their members (x) as ‘type’. The factor chosen as type is usually the most prominent, or best studied, one. Alternatively, the factor was chosen that was the first on alphabetical order when the family was defined. If there is only one member in a family, this one may be used to name the family, without the addition ‘-related’. If there are only two members in one family, they may be both mentioned in the family name. In case that all known members of a family share a biological function, which is also sufficiently discriminating against other clades, this function was preferentially used to name the family.
Subfamily names are similarly defined as family names. The subfamily containing the type of the whole family should be also named after this type, but as ‘x-like factors’ to differentiate against family names.
Numbering scheme
In analogy to the Enzyme Commission numbering system, the classification introduced here assigns a four-digit number to each TF that represents the top four classifying abstractions. It has been expanded by two more numbers for the physical entities assigned, so that each individual TF is unambiguously identified by a six-digit number. If not defined, optional levels are indicated by a ‘0’. The number ‘0’ may also indicate some uncertainty in the classification: it may well be that at some point of future developments, introduction of subfamilies may appear appropriate. For the same reason, some TFs have been assigned to a tenth superclass with the number ‘0’. These are TFs for which little knowledge exists about their DBD structure, although there may be experimental evidences that there is one; this domain may already be delineated in the molecule, which may have given rise to subsume some of them to a specific class, maybe even with some family structure underneath.
Obviously, any ‘0’ taxa are subject to change when the scientific insight expands. This is particularly true for Superclass ‘0’. However, it was introduced to reflect the present knowledge to the best extent possible, as limited it may be.
This will also facilitate to keep the numbering schema as stable as possible throughout all future developments. Optimally, the schema should be sufficiently robust so that any changes can be kept to a minimum, in particular to keep the numbers unchanged. That has been one of the main reasons to keep the order of the previous three superclasses (see also below, section Superclasses) as well as the order of classes and families to the utmost extent reasonable.
However, progress in future research may enforce major rearrangements, for instance to avoid huge gaps in the numbering scheme when large groups of previous TFs had to be deleted and where to be assigned at another place in the classification.
CONTENTS
Altogether, the TF classification presented here assigns 1558 human TF genes, encoding 2904 proteins (according to UniProt annotation), to 111 families, many of them being subdivided into subfamilies, of which 336 were defined. They constitute 40 classes and 10 superclasses (including the transitory Superclass ‘0’, ‘Yet undefined DNA-binding domains’), according to the features of their DBDs. The aim was to include all human transcriptional regulators that are known to bind to DNA in a sequence-specific manner, or that can be plausibly assumed to do so due to strong similarity with well-characterized TFs. Counting those TFs that have been experimentally proven to bind DNA in a sequence-specific manner and their closest homologs at the lowest classification level possible (subfamily or family) results in a total of 970 TF genes, or 62.3%, of the entries at genus level. The lowest percentage of proven DNA binders (28.4%) among the major classes is in the largest of them, the C2H2 zinc finger proteins.
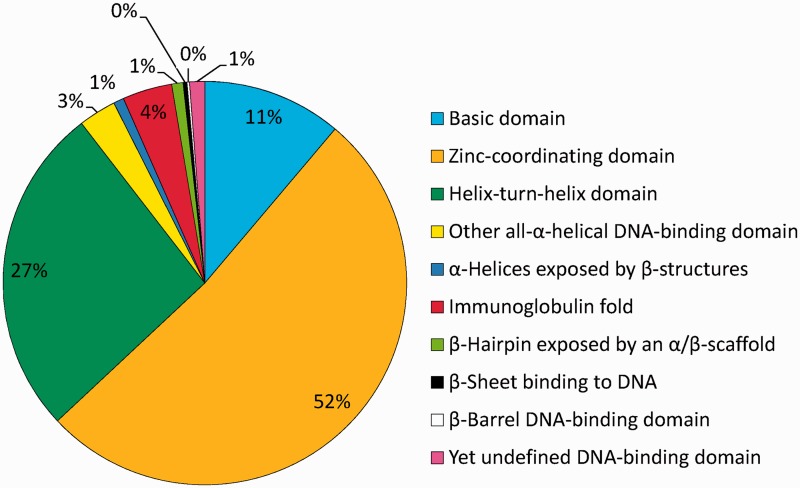
Among the nine superclasses of defined DBDs, by far the largest is Superclass 2 (zinc-coordinating DBDs) to which 53% of all TF genes belong to, followed by helix-turn-helix (26%) and basic domain factor genes (11%, Figure 1). These three superclasses have in common that an α-helix is exposed in such a way that it binds into the major groove of the DNA.
Figure 1.
Relative sizes of human TF superclasses. The distribution of TF genera among the 10 superclasses, with the percentages indicated.
Further statistics, e.g. about the size of individual taxa and the number of splice variants, are given in Supplementary Table S1.
Additional information
Additional information is provided for many taxa. For superclasses and classes, general descriptions are given, some of them were taken over from the TRANSFAC database and others were newly created, as is properly indicated.
Further information can be obtained for classes (except for those in Superclass 0) through separate HTML documents (link ‘More’ next to the rank term). Here, comprehensive alignments of the DBDs and their sequences in FASTA format are provided as well as logo plots for classes or, if appropriate, for their constituting families. In some cases, as for bZIP, basic helix-loop-helix (bHLH) or C4 zinc fingers (nuclear receptor type), dimerization matrices for the members of the corresponding class are provided for download. They give an overview about which class members are known to interact with each other, including the homodimers along the diagonal, illustrating the large variability of active agents some classes can generate. These matrices are downloadable as Excel sheets from the class information pages, which can be invoked from the link ‘more’ behind the rank in the window ‘Details’.
Most families, and sometimes subfamilies, have been assigned with one ‘typical’ DNA-recognition sequence. These sequences assigned to many families or subfamilies represent typical, idealized or proven individual binding sequences for (some of) the domains in the respective taxon. They should not be misunderstood as a consensus sequence, and they are not suitable for predicting potential TF-binding sites. Corresponding references are given as PubMed links; in some cases, they match with a consensus derived from a TRANSFAC positional weight matrix, as indicated in the Comment field.
Database cross-links
All ‘Genus’ entries have a link to the corresponding ‘UniProt’ entry and, whenever possible, to the corresponding entry in the TRANSFAC database. Entries at the molecular species level also show the corresponding UniProt isoform hyperlink and likewise connect to TRANSFAC. Through Ensembl gene numbers, ‘Genus’ entries are also associated with the protein expression signatures documented by the Human Protein Atlas (31).
IMPLEMENTATION
TFClass as ontology tree
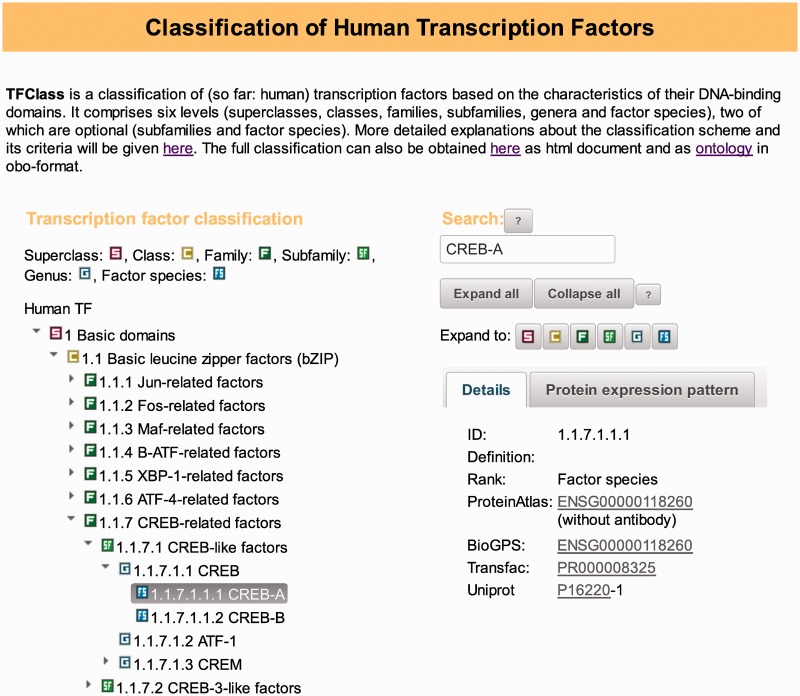
TFClass has been made available as an expandable tree, by default showing the topmost level, i.e. the superclasses. Individual branches can be expanded to the next level by clicking on the respective parent node (Figure 2). Besides, a fully expanded classification is available as HTML document as well.
Figure 2.
TFClass web interface. In the box on the left, the classification tree is shown and can be expanded by clicking on the individual items. In the example shown, the tree has been automatically expanded around the searched item (CREB-A, see ‘Search’ box in the top right part), and is cut after subfamily 1.1.7.2. The tree can also be browsed by expanding it down to any level (see area labeled ‘Expand to’). Under the tab ‘Details’ on the right, additional information about the active item is displayed directly or as link. Under the tab ‘Protein expression’, tissue specificities of the highlighted factor would be shown. On top, the ‘Search’ box enables to retrieve specific objects and open the tree on the left around the found items.
Clicking on the name of any taxon also invokes the additional information described above in a separate box headed ‘Details’, among them always the name and rank of the respective taxon. Further details for class members (except for those in Superclass 0) can be invoked through the link ‘More’ next to the rank term. From the page that will be reached through this link, the ‘Additional information’ described above is available. Under ‘Details’, cross-links to other databases are also depicted when clicking on a ‘Genus’ (or ‘Species’) entry.
Search function
The TFClass web interface provides a straight-forward search function. Any string entered is automatically extended by a wildcard. A list of search results is displayed, and selecting any item from this list and pressing the ‘GO’ button leads on an expansion of the classification tree around the matching entry.
CONCLUSION
The classification scheme of TFClass has been applied so far to human TFs, but has been designed in a way that it can be easily extended to other mammalian TFs without the requirement to adapt the structure. Later extension to other eukaryotic groups of organisms, in particular insects, plants and yeast, has already been anticipated as far as possible. One aim of the classification is to provide a systematic basis for assigning functional properties of TFs, first of all their DNA-binding characteristics. Similar DNA-binding capabilities at (sub-) family level can be exploited further for re-constructing extended transcriptional networks (Haubrock et al., submitted for publication). It will be interesting to see whether related factors occupy comparable positions in such networks. Systematic connection with expression patterns of the individual TFs, as done here by linking to Protein Atlas and soon to BioGPS as well, will help to construct tissue-specific transcriptional networks. A function will be implemented to retrieve TF signatures for individual tissues. In the long term, we will complement the classification by more functional criteria, which may give rise to orthogonal classifications. The present technical implementation as an ontological structure seems to be appropriate for the present status of the classification. It will be enriched with further search and visualization functions in near future.
AVAILABILITY
TFClass is freely accessible at http://tfclass.bioinf.med.uni-goettingen.de/ and has been made available in OBO format as a downloadable file. The latest draft is also available as a fully expanded HTML document at http://www.edgar-wingender.de/huTFclassification.html.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
The European Union’s Seventh Framework Programme for Research (FP7) (partly) [LipidomicNet: 202272, SysCol: 258236]; Federal Ministry of Education and Research (BMBF), Germany [GerontoShield: FKZ0315890B]. We acknowledge support by the German Research Foundation. Funding for open access charge: Göttingen University Medical School (UMG).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Olga Kel-Margoulis (GeneXplain GmbH) for her constant advice, and Volker Matys and Mathias Krull (BIOBASE GmbH) for their help in providing the links to the TRANSFAC database.
REFERENCES
- 1.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 2.Kel-Margoulis OV, Kel AE, Reuter I, Deineko IV, Wingender E. TRANSCompel: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res. 2002;30:332–334. doi: 10.1093/nar/30.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Hua X, Haubrock M, Wang J, Wingender E. The architecture of the gene regulatory networks for different somatic cells. Bioinformatics. 2012;28:i509–i514. doi: 10.1093/bioinformatics/bts387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki M, Yagi N. DNA recognition code of transcription factors in the helix-turn-helix, probe helix, hormone receptor, and zinc finger families. Proc. Natl Acad. Sci. USA. 1994;91:12357–12361. doi: 10.1073/pnas.91.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu WY, Huang YF, Huang CC, Cheng YS, Huang CK, OyangY J. ProteDNA: a sequence-based predictor of sequence-specific DNA-binding residues in transcription factors. Nucleic Acids Res. 2009;37:W396–W401. doi: 10.1093/nar/gkp449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison SC. A structural taxonomy of DNA-binding domains. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 9.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 10.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-reviews001. REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang HM, Chen H, Liu W, Liu H, Gong J, Wang H, Guo AY. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40:D144–D119. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulton DL, Sundararajan S, Badis G, Hughes TR, Wasserman WW, Roach JC, Sladek R. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 2009;10:R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wingender E. Compilation of transcription regulating proteins. Nucleic Acids Res. 1988;16:1879–1902. doi: 10.1093/nar/16.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 17.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wingender E. Classification of eukaryotic transcription factors. Mol. Biol. Engl. Tr. 1997;31:483–497. [PubMed] [Google Scholar]
- 20.Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 26.Hagopian R, Davidson JR, Datta RS, Samad B, Jarvis GR, Sjölander K. SATCHMO-JS: a webserver for simultaneous protein multiple sequence alignment and phylogenetic tree construction. Nucleic Acids Res. 2010;38:W29–W34. doi: 10.1093/nar/gkq298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dönitz J, Wingender E. The ontology-based answers (OBA) service: a connector for embedded usage of ontologies in applications. Front. Genet. 2012;3:197. doi: 10.3389/fgene.2012.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tipton K, Boyce S. History of the enzyme nomenclature system. Bioinformatics. 2000;16:34–40. doi: 10.1093/bioinformatics/16.1.34. [DOI] [PubMed] [Google Scholar]
- 29.Wingender E. Recognition of regulatory regions in genomic sequences. J. Biotechnol. 1994;35:273–280. doi: 10.1016/0168-1656(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 30.Knüppel R, Dietze P, Lehnberg W, Frech K, Wingender E. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol. 1994;1:191–198. doi: 10.1089/cmb.1994.1.191. [DOI] [PubMed] [Google Scholar]
- 31.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]