Abstract
Database annotations of prokaryotic genomes and eukaryotic mRNA sequences pay relatively low attention to frame transitions that disrupt protein-coding genes. Frame transitions (frameshifts) could be caused by sequencing errors or indel mutations inside protein-coding regions. Other observed frameshifts are related to recoding events (that evolved to control expression of some genes). Earlier, we have developed an algorithm and software program GeneTack for ab initio frameshift finding in intronless genes. Here, we describe a database (freely available at http://topaz.gatech.edu/GeneTack/db.html) containing genes with frameshifts (fs-genes) predicted by GeneTack. The database includes 206 991 fs-genes from 1106 complete prokaryotic genomes and 45 295 frameshifts predicted in mRNA sequences from 100 eukaryotic genomes. The whole set of fs-genes was grouped into clusters based on sequence similarity between fs-proteins (conceptually translated fs-genes), conservation of the frameshift position and frameshift direction (−1, +1). The fs-genes can be retrieved by similarity search to a given query sequence via a web interface, by fs-gene cluster browsing, etc. Clusters of fs-genes are characterized with respect to their likely origin, such as pseudogenization, phase variation, etc. The largest clusters contain fs-genes with programed frameshifts (related to recoding events).
INTRODUCTION
Frameshifts predicted by ab initio program GeneTack (1) correspond to reading frame transitions. The transition could be caused by many reasons, among them sequencing errors (2), indel mutations (3), programed frameshifting events (4–6), phase variation (7), overlapping of adjacent genes (8), dual-coding regions (9) and eukaryotic alternative splicing (10).
Although sequencing errors are artifacts of sequencing technologies, authentic indel mutations are features of real sequences. These mutations usually lead to gene pseudogenization; still some pseudogenization remain conserved in evolution if the transcript (not truncated contrary to the protein product) carry some function (11).
In case of phase variation, reversible indel mutations occur at high frequencies at specific sites. They generate a population of bacteria with heterogeneous sequences of phase variant gene, thus increasing population fitness, for example it may help some bacterial pathogens to escape immune response of a host (12). Phase variation results in reversible and inheritable variation of bacterial phenotype.
Programed frameshifting occurs either during translation (PRF, programed ribosomal frameshifting) or transcription (PTR, programed transcriptional realignment). PRF and PTR violate standard triplet decoding allowing for a single protein to be produced from two overlapping open reading frames (ORFs). Hence, GeneTack predicts frame transition between these ORFs. PRF and PTR occur at sites with specific sequence patterns conserved in evolution because programed frameshifting is required for gene expression. Programed frameshifting usually results in synthesis of two protein products (standard and frameshift) that share the same N-terminal sequence but possess different C-terminal parts. Among chromosomal genes, the best studied examples are bacterial prfB gene encoding Release Factor 2 (13) and eukaryotic genes encoding ornithine decarboxylase antizyme (14). PRF is abundant in viruses (15), bacteriophages and transposons (16,17). The largest available collection of known PRF genes is available in the Recode database (18).
A frameshift could be predicted when two coding sequences (CDSs) that are in different frames and located close to each other or overlap. Notably, a co-location of some of the CDS pairs could be evolutionary conserved if expression of the two genes is linked by translational coupling mechanism. Such gene pairs predicted as fs-genes are present in the GeneTack database as well.
Eukaryotic part of the database was built using known mRNA sequences; a large number of predicted fs-genes was found in alternative spliced transcripts containing premature termination codons (PTCs) (10,19). This fact is not surprising taking into account that in mammals upto one-third of alternative splicing (AS) events produce PTC-containing splice variants (20,21).
The database contains fs-genes that represent possible dual coding in eukaryotic mRNAs. Dual coding allows the same stretch of DNA to encode two protein sequences in different frames (22). Multiple instances of dual coding in human genome were detected by the analysis of ribosomal profiling data obtained from HeLa cells (23). Several instances of dual coding are well studied, such as the xbp1 gene encoding x-box binding protein 1. The products of initial rounds of xbp1 mRNA translation facilitate endonuclease-mediated excision of a 26-nt fragment of its own mRNA. As a result, mRNA downstream of excision appears in a different frame (24) and different protein product is synthesized from the same mRNA at the later rounds of translation. Another well-studied example (9) is expression from the GNAS1 locus (coding for Guanine Nucleotide binding protein Alpha Subunit 1) an alternative protein called ALEX (“ALternative gene product Encoded by XL-exon”). Similarly, tumor suppressor proteins P16(INK4a) and P14(ARF) are produced from the same gene, where the same sequence appears in alternative frames in two alternative transcripts (25). Due to the codon co-dependency of overlapping frames (26) dual coding regions have unusual codon frequencies that make them prone to frameshift prediction by GeneTack.
The GeneTack database contains all types of frame transition events (prokaryotic and eukaryotic); ∼20% of the entries have been characterized in terms of the probable nature of predicted frame transition.
To help explore the nature of predicted fs-genes, they were grouped into clusters of orthologous fs-genes based on sequence similarity, conservation of frameshift direction (−1, +1) and location. We characterized the fs-genes that formed the largest clusters based on comparative genomics analysis (Antonov I et al., submitted for publication). Although the nature of >80% of the predicted frameshifts was not revealed (at least 1.5% have a strong evidence to be sequencing errors, whereas upto 54% could be related to sequencing errors), this database will be useful for improving annotation of new genomes, re-annotation of old ones as well as for stimulating experimental studies leading to identification of new programed events and other cases of frame transitions under evolutionary selection.
DATABASE STATISTICS AND USAGE
The data are stored in a local MySQL database queried by CGI scripts embedded in the web interface. The database also includes some pre-built data, such as Sequence LOGOs (27) of conserved motifs observed in overlapping ORFs for all the clusters.
The database consists of two sections—prokaryotic and eukaryotic. Notably, the method of frameshift prediction was slightly different in prokaryotic genomic DNA and eukaryotic mRNA. For prokaryotes, genes in a complete genome sequence were predicted by GeneMarkS (28), the self-training program that derived parameters both for itself, as well as for GeneTack. A single statistical model was generated for each prokaryotic genome and use in GeneTack.
Eukaryotic genes with frameshifts were identified in mature mRNA sequences. Several HMM models were generated for each eukaryotic genus. Each model was generated by a self-training algorithm, a version of GeneMarkS, from a set of mRNAs with a close GC percent content. All the eukaryotic and prokaryotic models are available at the GeneTack web page; a database user can choose an appropriate pre-built model for a query sequence.
Currently, the database contains fs-genes from 1106 prokaryotic and 100 eukaryotic species (Table 1). Since the length of prokaryotic genomes as well as the total size of available eukaryotic mRNAs vary for different species, the number of predicted fs-genes also varies. For example, in 115 001 human mRNAs, we predicted 8700 frameshifts, whereas only 839 frameshifts were predicted in 32 155 mRNA sequences of Rattus norvegicus. Conceptual translation of predicted fs-genes produced a database of fs-proteins used for clustering. All over, 50% of prokaryotic and 27% of eukaryotic fs-genes formed clusters, whereas other fs-genes were singletons.
Table 1.
Statistics on eukaryotic and prokaryotic sections of the GeneTack database
| Database statistics | Prokaryotes | Eukaryotes |
|---|---|---|
| Number of species analyzed | 1106 | 100 |
| Total number of predicted frameshifts | 206 991 | 45 295 |
| Total number of clusters | 19 430 | 4087 |
| Number of fs-genes in all clusters | 102 731 | 12 103 |
| Number of singleton fs-genes | 104 260 | 33 192 |
| Number of clusters with less than five fs-genes | 14 441 | 3701 |
| Number of programmed frameshift clusters | 146 | 5 |
| Number of indel mutation clusters | 4010 | 2 |
| Number of clusters of PTC-containing splice variants | n/a | 21 |
The database home page is the user’s entry point. The user can browse prokaryotic or eukaryotic clusters of fs-genes, perform sequence similarity search by BLASTp for a query sequence of interest or search for fs-genes or clusters using a query string. The query string could be fs-gene/cluster identification number (ID) or cluster name.
Majority of the clusters were named using names of Pfam domain detected in the cluster of fs-proteins. However, several clusters (e.g. known cases of programed frameshifting) were manually renamed to reflect gene and protein names. Thus, Release Factor 2 cluster can be found by using the gene name ‘prfB’ as a query.
To allow search against the GeneTack database of fs-proteins, two BLASTp databases (containing either prokaryotic or eukaryotic fs-proteins) were built. The BLASTp hit may reveal the nature of a frameshift mechanism in a novel sequence.
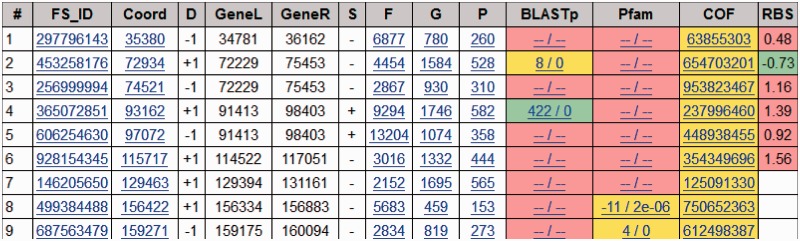
Finally, a user can browse sections of either of the two databases in the following ways. First, a particular species can be selected from a list of species. For a given species, a list of all the predicted fs-genes is available (Figure 1). The list provides information about every frameshift such as its direction and genomic coordinates. More detailed information about an fs-gene can be accessed by clicking on the fs-gene ID. A page with frameshift details provides the following information: the species name, the frameshift coordinate (in the prokaryotic genome or the eukaryotic mRNA), the frameshift direction (+1 or −1), the coordinates of the fs-gene, its length and the length of encoded protein. The initial fs-gene sequence (with a frameshift), the corrected fs-gene sequence and the sequence of conceptually translated protein product are available as well. Additional information for a frameshift includes reference to the BLASTp/Pfam hit if it did occur to cover predicted frameshift position in the fs-protein. Link to the corresponding cluster is provided if the fs-gene belongs to the cluster. It should be noted that an fs-gene can belong to one cluster only.
Figure 1.
The GeneTack database entries: fs-genes predicted in the genome of Escherichia coli str. K-12 substr. DH10B. FS_ID—unique fs-gene identificator, Coord—frameshift coordinate in the input sequence, D—frameshift direction (+1 or −1), GeneL coordinate of left border of fs-gene (gene start for ‘+’ strand, gene end for ‘−’ strand), GeneR—coordinate of right border of fs-gene (gene end for ‘+’ strand, gene start for ‘−’ strand), S—the fs-gene strand, F—frameshift coordinate in fragment (the sequence used as input to GeneTack), G—frameshift coordinate in fs-gene, P—frameshift coordinate in fs-protein, BLASTp—information on the BLASTp hit covering frameshift position in the fs-protein, Pfam—information on the Pfam domain covering frameshift position in the fs-protein, COF—cluster ID (if available), RBS—RBS score of the downstream gene defined by GeneMarkS.
Another way of browsing the database is by using a probable type of fs-gene. Some of the predicted fs-genes and the clusters of the fs-genes were grouped together based on their types. Each group of the clusters (for example, all prokaryotic programed frameshift clusters) can be seen as a list on a single web page with general information about each cluster.
The type of a cluster was predicted using a range of cluster’s gene features. To identify programed frameshift clusters, sequences in the vicinities of the frameshifts were analyzed in order to find a conserved motif that would resemble a frameshift site. Protein products from pseudogene clusters must have BLASTp hits in nr database indicating that predicted frameshift is a result of an indel mutation. Elevated frequency of tandem repeats near predicted frameshifts was chosen as a characteristic property of a phase variation clusters. On the other hand, conserved start codons for downstream ORF2 are expected in the vicinity of the frameshifts in translational coupling clusters.
There are a number of large clusters for which the nature was not predicted but they may be of interest to research community. To provide access to these clusters, additional groups were introduced: clusters with 100+ and 50–100 fs-genes (in case of prokaryotes) and 10+ fs-genes (in case of eukaryotes), so clusters could be retrieved by size.
Additionally, during the search for prokaryotic programed frameshift clusters, we have analyzed the frameshift vicinities and grouped clusters by the most over-represented heptamer. The heptamers include special symbols (underscores) to indicate the reading frame of the upstream ORF1.
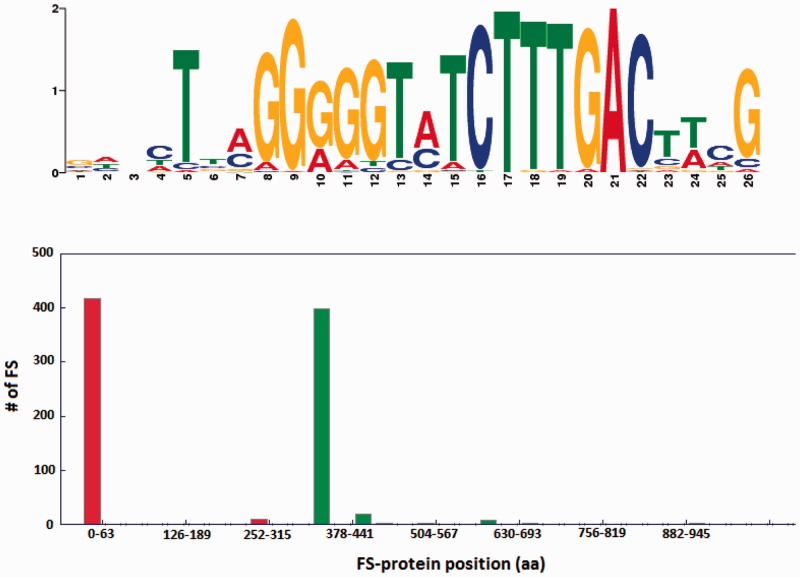
The cluster details page contains the same information as the fs-gene details page except that the information is provided for all the clusters’ fs-genes together, e.g. a multi-fasta file where all the fs-gene or the fs-protein sequences are provided instead of a single sequence. The cluster information page may also include figures visualizing frequencies of nucleotides in conserved motifs (sequence LOGOs) located close to the frameshift position, as well as the distributions of frameshift coordinates and the fs-gene lengths (Figure 2). Sequence LOGOs were generated with the MEME software package (29).
Figure 2.
Logo of the conserved motif (upper panel) and distribution of coordinates of frameshifts (lower panel) in 428 fs-genes of Release Factor 2 collected in a cluster (ID 474411093) (13). Red bars in the lower panel correspond to frameshift positions and green bars show the total length of fs-proteins. The small green bars indicate existence of subgroups of longer fs-proteins.
TOOLS FOR FRAMESHIFT PREDICTION
Besides the database, the GeneTack server contains a number of tools for frameshift identification in nucleotide sequences. There are four main programs—GeneTack-GM (1), GeneTack-Prok (1), GeneTack-Euk (Antonov I et al., manuscript in preparation) and MetaGeneTack (Tang S et al., accepted for publication to Bioinformatics).
GeneTack-GM is a combination of frameshift prediction program GeneTack and a self-training gene prediction program GeneMarkS (28). GeneTack-GM could be used to predict frameshfits in long prokaryotic sequences (longer than 300 kB). The model parameters are automatically generated by a self-training program GeneMarkS. GeneTack-GM also includes a number of filters to remove false-positive predictions.
GeneTack-Prok and GeneTack-Euk can be used to analyze shorter prokaryotic and eukaryotic sequences with length insufficient for self-training. Eukaryotic sequences must be intronless, e.g. mRNAs or expression sequence tags (ESTs) can be used. Both programs feature a number of pre-built species-specific models. A user should choose the one that corresponds to the input sequence. No filters are applied to the frameshifts predicted by these two programs.
GeneTack cannot be directly applied to short metagenomic sequences because it requires a species-specific statistical model. Yet another ab initio frameshift finder, MetaGeneTack, can be used in this case (Tang S et al., accepted for publication). MetaGeneTack uses heuristic models (30) and applies several additional filters for removing false-positive predictions.
APPLICATION OF THE TOOLS AND DATABASE
The GeneTack tools predict frameshifts in all types of sequences. Using one of the tools, a user can find candidate genes with frameshfits in a new prokaryotic genome, contig or metagenome or explore a single protein-coding mRNA for a presence of frameshifts. The predicted fs-genes are automatically translated into fs-proteins that could be used as queries against GeneTack database. Hits to large clusters will show phylogenetic conservation of the frameshift. An association with a large cluster can be used to argue that the predicted frameshift is not a result of sequencing error. Moreover, if the type of the cluster is known (e.g. programed frameshift) it is likely that the input sequence has a frameshift of the same type as well.
AVAILABILITY
The interface to GeneTack database is at http://topaz.gatech.edu/GeneTack/db.html. All data are available for download as flat files (sequences in fasta format) and also as a set of MySQL relational database files. Each fs-gene as well as each fs-gene cluster has a unique ID. The genes or clusters are accessible through URLs: http://topaz.gatech.edu/GeneTack/cgi/fs_view.cgi?id=FS_ID (for fs-genes) or cof_view.cgi?id=CLUSTER_ID (for clusters).
FUNDING
Funding for open access charge: The work of I.A. and M.B. was supported in part by the USA National Institute of Health grant [HG000783 to M.B.]; the work of P.V.B. was supported in part by Wellcome Trust grant [094423 to P.V.B.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Alexandre Lomsadze for valuable help.
REFERENCES
- 1.Antonov I, Borodovsky M. GeneTack: frameshift identification in protein-coding sequences by the Viterbi algorithm. J. Bioinformatics Comput. Biol. 2010;8:535. doi: 10.1142/s0219720010004847. [DOI] [PubMed] [Google Scholar]
- 2.Medigue C, Rose M, Viari A, Danchin A. Detecting and analyzing DNA sequencing errors: toward a higher quality of the Bacillus subtilis genome sequence. Genome Res. 1999;9:1116–1127. doi: 10.1101/gr.9.11.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshayes C, Perrodou E, Gallien S, Euphrasie D, Schaeffer C, Van-Dorsselaer A, Poch O, Lecompte O, Reyrat JM. Interrupted coding sequences in Mycobacterium smegmatis: authentic mutations or sequencing errors? Genome Biol. 2007;8:R20. doi: 10.1186/gb-2007-8-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 5.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 6.Dinman J. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip. Rev. RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Woude MW. Re-examining the role and random nature of phase variation. FEMS Microbiol. Lett. 2006;254:190–197. doi: 10.1111/j.1574-6968.2005.00038.x. [DOI] [PubMed] [Google Scholar]
- 8.Pradhan P, Li W, Kaur P. Translational coupling controls expression and function of the DrrAB drug efflux pump. J. Mol. Biol. 2009;385:831–842. doi: 10.1016/j.jmb.2008.11.027. [DOI] [PubMed] [Google Scholar]
- 9.Klemke M, Kehlenbach RH, Huttner WB. Two overlapping reading frames in a single exon encode interacting proteins–a novel way of gene usage. EMBO J. 2001;20:3849–3860. doi: 10.1093/emboj/20.14.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C, Krainer AR, Zhang MQ. Evolutionary impact of limited splicing fidelity in mammalian genes. Trends Genet. 2007;23:484–488. doi: 10.1016/j.tig.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Khachane A, Harrison P. Assessing the genomic evidence for conserved transcribed pseudogenes under selection. BMC Genomics. 2009;10:435. doi: 10.1186/1471-2164-10-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 14.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firth A, Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93(Pt 7):1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranov P, Fayet O, Hendrix R, Atkins J. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Sharma V, Firth A, Antonov I, Fayet O, Atkins J, Borodovsky M, Baranov P. A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. Mol. Biol. Evol. 2011;28:3195–3211. doi: 10.1093/molbev/msr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bekaert M, Firth AE, Zhang Y, Gladyshev VN, Atkins JF, Baranov PV. Recode-2: new design, new search tools, and many more genes. Nucleic Acids Res. 2010;38:D69–D74. doi: 10.1093/nar/gkp788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGlincy N, Smith C. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Q, Saltzman A, Kim Y, Misquitta C, Shai O, Maquat L, Frey B, Blencowe B. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung W, Wadhawan S, Szklarczyk R, Pond S, Nekrutenko A. A first look at ARFome: dual-coding genes in mammalian genomes. PLoS Comput. Biol. 2007;3:e91. doi: 10.1371/journal.pcbi.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel AM, Roy Choudhury K, Firth AE, Ingolia NT, Atkins JF, Baranov PV. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012;22:2219–2229. doi: 10.1101/gr.133249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 25.Ouelle D, Zindy F, Ashmun R, Sherr C. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 26.Nekrutenko A, Wadhawan S, Goetting-Minesky P, Makova K. Oscillating evolution of a mammalian locus with overlapping reading frames: an XLαs/ALEX relay. PLoS Genet. 2005;1:e18. doi: 10.1371/journal.pgen.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider T, Stephens R. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]