Abstract
We present a database of repetitive DNA elements, called Dfam (http://dfam.janelia.org). Many genomes contain a large fraction of repetitive DNA, much of which is made up of remnants of transposable elements (TEs). Accurate annotation of TEs enables research into their biology and can shed light on the evolutionary processes that shape genomes. Identification and masking of TEs can also greatly simplify many downstream genome annotation and sequence analysis tasks. The commonly used TE annotation tools RepeatMasker and Censor depend on sequence homology search tools such as cross_match and BLAST variants, as well as Repbase, a collection of known TE families each represented by a single consensus sequence. Dfam contains entries corresponding to all Repbase TE entries for which instances have been found in the human genome. Each Dfam entry is represented by a profile hidden Markov model, built from alignments generated using RepeatMasker and Repbase. When used in conjunction with the hidden Markov model search tool nhmmer, Dfam produces a 2.9% increase in coverage over consensus sequence search methods on a large human benchmark, while maintaining low false discovery rates, and coverage of the full human genome is 54.5%. The website provides a collection of tools and data views to support improved TE curation and annotation efforts. Dfam is also available for download in flat file format or in the form of MySQL table dumps.
INTRODUCTION
Copies of transposable elements (TEs) at various levels of decay make up a large fraction of many genomes in the form of interspersed repetitive DNA. Accurate annotation of TEs enables research into their fascinating biology, impact on the genome of the host organism and the evolutionary processes that shape genomes. Most TE annotations are performed using RepeatMasker (http://www.repeatmasker.org) or Censor (1), both of which depend on the Repbase database of repetitive DNA elements (2,3). The sensitivity of TE detection depends on both database content and homology search method. In the Repbase database, each TE entry is represented by a single consensus sequence. RepeatMasker and Censor depend on a variety of homology search tools — cross_match (http://www.phrap.org), rmblastn (http://www.repeatmasker.org/RMBlast.html) and abblast (http://blast.advbiocomp.com) — each of which searches for pairwise similarity between a sequence of interest and the collection of all consensus sequences. Although these methods annotate TEs covering substantial portions of many genomes (current coverage for human is 51.3%, see http://www.repeatmasker.org/species/homSap.html), this is expected to be incomplete because older TE instances may not be recognized as a result of extensive mutation. Profile methods represent an entry using an alignment of multiple representative sequences rather than a single consensus and are known to improve sensitivity over single sequence search (4), with profile hidden Markov models [profile HMMs (5)] in particular leveraging the additional information content in position-specific residue and indel (insertion and deletion) variability.
To date, it has not been possible to apply profile HMM search to TE annotation because DNA search was too slow. However, the new heuristic filtering pipeline and efficient vector-parallel implementation of HMMER3 (6) provide the foundation for a new tool, nhmmer (to be described in detail elsewhere), which brings the power of profile HMMs to DNA homology search with good speed (still slower than a sensitive parameterization of blastn, but faster than cross_match with sensitive parameters).
We present Dfam, a database of curated high-quality profile HMMs for all TEs known in the human genome. Dfam is the product of collaboration between the developers of Repbase, RepeatMasker, HMMER and the Xfam consortium (7,8). The program nhmmer has been incorporated as a search engine for RepeatMasker, and the Dfam HMM library can be used by RepeatMasker to annotate TEs in the human genome.
In our tests, the combination of Dfam and nhmmer produces annotation of an additional 2.9% of a large sample of the human genome (a 516-Mb benchmark from the human genome, GRCh37.p7 assembly) over cross_match with the Repbase RepeatMasker library, without increasing false discovery rate or sacrificing speed. The Dfam website (http://dfam.janelia.org) enables searching of an uploaded sequence against the HMM library and provides insight into the construction, sensitivity, relationships and distribution characteristics of each entry. Dfam is initially focused on human TEs because there are many well-studied, old elements and because gains in sensitivity will enable improved annotation of the human genome, which is arguably the most important genome for TE annotation. Over time, the Dfam library will grow to include TE entries for other organisms, via a combination of building on the mature collection of TE families in Repbase, and providing curation tools to facilitate the acquisition of new entries from the wider scientific community.
DESCRIPTION OF DFAM
The current release of Dfam, version 1.1, contains entries representing all TEs identified in the human genome. The Dfam database shares many design principles with Pfam and Rfam. Each entry is represented by a multiple sequence alignment, a profile HMM, curated entry-specific score thresholds and a listing of the location of nhmmer-identified matches to that entry in the human genome. In total, Dfam 1.1 contains 1143 entries: 767 retrotransposons, 240 DNA transposons, 28 interspersed repeats of unknown origin and 108 non-TE entries used to annotate satellites (35 entries) or to avoid annotating non-coding RNA genes (73 entries) as TEs.
The entries in Dfam are intended to be a drop-in replacement for the Repbase-derived RepeatMasker library of consensus sequences used by RepeatMasker for repeat detection in the human genome. The names of these models will not always match the final RepeatMasker annotation. One cause is that many complete TEs are broken into multiple Dfam entries representing a portion of the TE. For example, a full-length L1 retrotransposon can be broken into three entries: 5′-end, ORF 2 and 3′-end. RepeatMasker then makes complex conversions from entry names to final annotation; the result is not always immediately intuitive. For example, RepeatMasker maps adjacent occurrences of the L1M5_5end and L1ME3C_3end models to a final annotation of L1ME3C. Thus, the mapping from simple Dfam annotation to RepeatMasker annotation will not be perfect. We plan to make these mappings explicit in a future Dfam release.
SENSITIVITY AND FALSE DISCOVERY
To assess the utility of using Dfam for TE annotation, we tested sensitivity and false discovery rate (FDR). All models were searched against human chromosomal sequence (see later for details), and a base was called ‘covered’ if it was part of at least one search match.
A common practice in homology search is to use simulated sequence, for example preserving dinucleotide frequency, to estimate false positive (FP) rates. In tests on a benchmark made up of dinucleotide-preserved sequence, we found that the Dfam+nhmmer FP rate was remarkably low, and failed to highlight obvious simple repeat hotspots observed in a few models on genomic sequence (data not shown; for discussion of these hotspots, see the ‘Model Masking and Thresholds’ section). This is not surprising, as sequences produced in this way are homogenized, and do not contain all the typically problematic features of real genomic sequence like simple repeats and other low complexity regions such as poly-pyrimidine runs. On the other hand, reversed (but not complemented) genomic sequence does contain these features while theoretically removing true TEs from the sequence. As expected, searching against reversed genomic sequence corroborated simple repeat hotspots. There are legitimate concerns about k-mer composition discrepancies between chromosomal and reversed sequence, but FP estimates from using reversed genomic sequence seem to be reasonably accurate: in each case that a model was masked to hide a simple repeat hotspot, the numbers of hits lost on genomic and reversed sequence were typically similar (within 50% of the same count).
In practice, some TE sequences show similarity to their reversed sequences because of patterns of low complexity. To account for this, a match to reversed sequence was ignored (called a ‘neutral match’) if it corresponded to the location of a longer and higher-scoring match to the same model in the non-reversed sequence. Remaining matches were called ‘false matches’, and were used to compute false coverage using the same method as with chromosomal coverage. This false coverage was assumed to approximate the amount of false coverage on the actual chromosomal sequence, so that we defined FP and true positive (TP) values in units of nucleotides covered as:
and estimated the FDR within the chromosomal coverage as:
Specifically, human chromosomes 1, 2 and 19 (GRCh37.p7) were divided into adjacent non-overlapping blocks of 60 000 bases. Blocks with >10% N’s were removed from the data set, leaving 516.2 Mb in 8604 blocks, in which simple tandem repeats were masked using TRF (9) (with parameters ‘2 7 7 80 10 70 5 –d –h –m’). Chromosomes were divided in this way to test the impact of GC content on sensitivity and false discovery; no data are shown regarding GC impact, as GC had little impact on relative efficacy of tested methods.
The profile HMM approach of using Dfam + nhmmer was compared with alternative search tools cross_match (v.0.990329), NCBI blastn (2.2.25+) and rmblastn (2.2.23+). The program cross_match is the tool used by default in RepeatMasker because it gives the highest sensitivity. The program rmblastn is a variant of NCBI blastn optimized for TE search in RepeatMasker. Parameters for these two tools were chosen as the best single parameterization used by RepeatMasker (Table 1), based on our experience using these tools. To show that cross_match and rmblastn give substantially better results than naïve search, we include blastn results with two variants, one using default settings and one using the best performing settings among many alternatives we tested. The search library used for these tools consisted of the Repbase/RepeatMasker consensus sequences corresponding to each Dfam model.
Table 1.
Coverage and false discovery on benchmark data
| Method | Covered bases | Covered (%) | FP bases | FP (%) | FDR (%) | Time (h) |
|---|---|---|---|---|---|---|
| nhmmer | 278 140 893 | 53.88 | 159 028 | 0.03 | 0.06 | 595 |
| cross_match | 263 131 978 | 50.97 | 282 672 | 0.05 | 0.11 | 2682 |
| rmblastn | 257 212 437 | 49.82 | 201 430 | 0.04 | 0.08 | 59 |
| blastn (sensitive) | 231 296 716 | 44.80 | 135 832 | 0.03 | 0.06 | 28 |
| blastn | 201 836 787 | 39.10 | 68 743 | 0.01 | 0.03 | 18 |
Covered bases were computed by running a search of each entry model or consensus sequence against a 516.2-Mb benchmark from human chromosomes 1,2 and 19. FP nucleotides were computed as described in the text. FDR is the ratio of FP nucleotides to covered nucleotides. The software nhmmer (version snap-10162012) was run with default parameters, after building models using the flags (–hand –maxinsertlen 10) to ensure one match state for each position in the consensus, and to limit insert length parameterization, respectively. The software cross_match (v.0.990329) was run using RepeatMasker parameters calculated to be optimal for copies 25% diverged from their original sequence and in a background of 41% GC DNA (-gap_init -25 -gap_ext -5 -minmatch 7 -bandwidth 14 -masklevel 10 -matrix 25p41g.matrix –minscore 200). The software rmblastn (2.2.23+) was run with parameters that mirror those of cross_match, (-gapopen 20 -gapextend 5 -complexity_adjust -word_size 7 -xdrop_ungap 400 -xdrop_gap_final 800 -xdrop_gap 100 -min_raw_gapped_score 200 -dust no -matrix 25p41g.matrix). The software blastn (2.2.25+) was run with basic settings (-wordsize 7) and with sensitive settings (-reward 1 -penalty -1 -gapopen 2 -gapextend 1 -wordsize 7). For all tools, entry-specific score thresholds were chosen to meet a target FDR of 0.2%, as described in the text. Runtime was collected on a single thread on a 2.66 GHz Intel Gainestown (X5550) processor. Results show that the speed of nhmmer lies between that of rmblastn and cross_match.
For all tools, entry-specific score thresholds were established for each Dfam entry as follows. Each model was searched against both genomic and reversed sequence. A threshold was set for each entry as the lowest score at which the empirical FDR for hits at or above that score was less than the target FDR of 0.2%. This conservative target FDR was chosen because it matches the estimated FDR of RepeatMasker on human sequence, and ensures that annotation based on Dfam is reliable. The threshold was also required to be higher than all but the 10 highest-scoring false hits, and at least as high as the score corresponding to an E-value of 20. These secondary thresholds were chosen as simple methods of restricting hits with unreasonably liberal scores, which will otherwise be allowed for entries showing many thousands of genomic hits.
Table 1 shows results for these various tools. Without sacrificing FDR, the combination of Dfam and nhmmer produces an additional 2.9% coverage beyond that achieved using cross_match or rmblastn. Roughly half of the gain in coverage is because of new instances (hits found by nhmmer but missed by the other tools), and half because of extension of shared instances. Approximately 49% of new instances belong to the superfamilies L2 and MIR (32 and 17%, respectively), which account for only 23% of all nhmmer hits (11 and 12%, respectively) in this benchmark. A table of coverage and FDRs on a per-family basis is available in supplementary material (see the ‘Availability’ section).
Table 1 is a simplification of a broader survey we have performed regarding sensitivity, and suggests at least two concerns regarding tool comparison, which we address here. (i) The levels of FDR observed in Table 1 disagree with the per-family FDR target in various ways as a result of opposing forces: (a) the E-value and maximum FP constraints reduce the empirical FDR for families with many matches (e.g. Alu entries have >1 million matches, so 2000 FPs per Alu entry would be required to reach FDR = 0.2%, but not >10 are allowed); and (b) many entries show substantial redundancy in genomic matches but not in false hits, causing false covered base counts to grow more quickly than true covered base counts as additional Dfam entries are considered. (ii) The tool blastn shows lower FP levels than the other tools under the particular constraints of this experiment. This suggests the possibility that the relatively low sensitivity of blastn is simply the result of overly stringent parameterization, but this is not the case. We have tested numerous alternative blastn parameters, both in terms of runtime arguments and family threshold methods, and have found no variant that exceeds 235 million covered bases with fewer than 500 000 FP bases.
Overextension
The aforementioned reverse coverage test can be used to assess the rate and coverage of de novo false hits, but another possible source of false coverage is so-called ‘homologous overextension’ (10), the extension of a hit beyond the true bounds of a true instance and into flanking non-homologous sequence. Consider a region of sequence matched by multiple tools: all tools are expected to match this region in a repeated search, and some tools might extend into flanking sequence; the concern is establishing whether that extension is legitimate (a result of better sensitivity to the signal of true flanking match) or not (a result of overly permissive extension of hits).
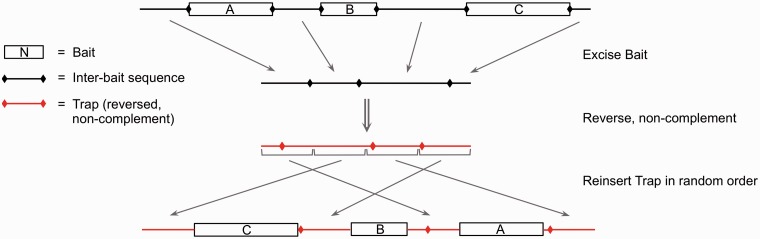
To test for overextension, we embedded trusted TE sequences into a chromosomal background expected to lack TEs, then searched with all models to identify covered bases as indicated earlier. The number of covered background positions is an estimate of overextension. Specifically, we defined a conservative set of TP TE subsequences by identifying the intersection of nucleotides matched by nhmmer, cross_match and rmblastn. Each contiguous run of these covered bases was called a ‘bait sequence’. If two contiguous bait sequences are partial instances of a long TE and are placed into background sequence in their original order, an alignment tool may extend through the background and (arguably reasonably) connect the partial instances into a longer hit. To avoid this, the order of bait sequences within each 60-kb block was inverted, as shown in Figure 1. Within each 60-kb block, all inter-bait sequences were concatenated and reversed (without complementation), and then divided into equal sized blocks. These were then placed in random order between the bait sequences (Figure 1). The sequence now flanking each TE bait was considered a ‘trap’. These traps broadly match the overall GC content of the original context of the bait sequences. We then measured each method’s tendency to overextend into flanking sequence (trap) seeded from true matches (bait).
Figure 1.
Construction of the overextension trap. Bait sequences (a conservative set of bases matched by nhmmer + Dfam and both cross_match and rmblastn with consensus sequences) were placed in inverted order. Inter-bait sequences were concatenated into a long stretch of sequence that was reversed without complementation and divided into equal sized blocks, which were then placed in random order between the bait sequences.
The results in Table 2 show that Dfam’s improved sensitivity does not come at the cost of false overextension. In this test, Dfam + nhmmer shows lower tendency to extend beyond the bounds of the bait sequence. The rate of overextension shown for all tools (∼3%) is a pessimistic estimate of improper coverage in a practical genome annotation: (i) in this test, each bait is separated by a long trap, while TE instances are frequently adjacent to each other in real genomic sequence, which makes overextension impossible; and (ii) many overextensions in this test are reasonable outcomes, for example, when the poly-A tail of an Alu model extends into an A-rich segment of trap adjacent to an Alu bait. We note that early tests of Dfam + nhmmer showed overextension to be a significant problem, especially in regions of high composition bias; the results in Table 2 are the outcome of improvements to nhmmer in response to those tests.
Table 2.
False coverage by overextension
| Tool | Bait covered (bp) | Trap covered (bp) | FDR (%) | Fraction of trap hit (%) |
|---|---|---|---|---|
| nhmmer | 244 069 604 | 6 200 737 | 2.48 | 2.30 |
| cross_match | 244 161 363 | 7 704 614 | 3.06 | 2.85 |
| rmblastn | 244 351 772 | 8 483 688 | 3.36 | 3.14 |
‘Bait’ and ‘trap’ sequences were produced as described in Figure 1 and the text. Total size of this benchmark was 516.2 Mb, of which 246.1 Mb was bait and 270.1 Mb was trap. FDR is defined as Trap bases/(Bait bases + Trap bases).
RELATED TOOLS
The Dfam database is supported by a collection of new tools that will appear in the future release of HMMER3.1. A release snapshot of HMMER3.1, including the version of nhmmer used to produce the database and the results in this article, is available via the FTP link at the top of every Dfam web page. In addition, the upcoming release of RepeatMasker (version 4.0) will incorporate Dfam and nhmmer (http://repeatmasker.org).
DFAM ENTRY CURATION PROCESS
The starting point for each entry in Dfam is the generation of (i) the seed alignment, a multiple alignment of representative sequences; and (ii) the profile HMM based on that alignment. The profile HMM characterizes the positional variability in both residue conservation and indel rates observed in the alignment, providing additional search power compared with a single consensus sequence and a uniform scoring model. Alignments and profile HMMs were produced as follows.
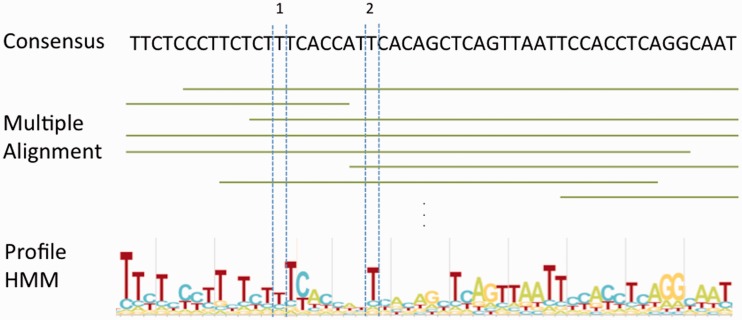
Multiple alignment tools like MUSCLE (11) are unable to create reasonable alignments for sequences with the level of divergence observed in most TE families. We therefore aimed to leverage the substantial effort that has gone into defining consensus sequences for the numerous TE subfamilies (most of which have been reconstructed from long inactive relics). We have built a multiple sequence alignment for each Dfam entry based on these consensus sequences, rather than starting from scratch. To produce the seed alignment for a Dfam entry, up to 2000 instances of the TE were semi-randomly selected from the output of a RepeatMasker analysis of the human reference genome as follows. RepeatMasker was run using cross_match at maximum sensitivity with the Repbase RepeatMasker library of consensus sequences. If >2000 TE instances were available, those with alignments covering more than 75% of the consensus length were preferentially used. If necessary, this set was supplemented with a selection of shorter instances to achieve (if possible) at least 10× coverage at each position in the consensus. Any instances in the upper quartile of the divergence range against the consensus were ignored to avoid inclusion of distant matches against related but as yet undescribed TEs, which could dilute the signal in the alignment (with increased divergence comes an increase in risk of incorrect alignment, which produces noise in the columns containing misaligned bases; removing these distant instances resulted in slightly better benchmark performance). The alignments against the consensus produced by RepeatMasker were merged into a multiple sequence alignment, as represented in Figure 2.
Figure 2.
Schematic representation of the creation of the multiple sequence alignment and profile HMM for a Dfam entry. The consensus and HMM logo correspond to positions 253–304 of Tigger16a (DF0000028), and highlight the difference between the abilities of HMM and consensus to represent positional residue conservation — a consensus treats all majority rule decisions as equivalent, while a profile HMM enables position-specific scoring based on conservation. In this case, the position labelled with (1) has a slight preference for ‘T’, but will not substantially reward a ‘T’ or penalize any other nucleotide; meanwhile the position labelled with (2) shows a strong preference for ‘T’, and will provide high reward for a matching ‘T’, and a strong penalty for any other nucleotide.
As in Pfam (7) and Rfam (8), we maintain an underlying primary sequence database, with an assigned version, so that results of each release can be reliably reproduced. This database, called dfamseq, currently consists of just the human genome (human assembly as downloaded from Ensembl, release 67, corresponding to GRCh37.p7, including non-placed contigs, but excluding the mitochondrial DNA). In the future, dfamseq will grow to include many more genomes. All sequences in current seed alignments are of human origin, and found in dfamseq.
Model masking and thresholds
A profile HMM for each entry was produced from the seed alignment using the HMMER tool hmmbuild. We ensured that the model contains a match state position for each nucleotide in the original consensus by including a custom RF line in the seed alignment, and running hmmbuild with the ‘–hand’ flag. Position-specific expected insert length was restricted to no longer than 10 with ‘–maxinsertlen 10’, as long seed alignment inserts can otherwise induce nhmmer to match unacceptably long inserts bridging legitimate partial hits. Dfamseq was masked for simple tandem repeats using the program Tandem Repeats Finder (TRF, 9), and then reversed (without complementation) to form dfamseq-rev. Models were searched against dfamseq and dfamseq-rev using nhmmer with an E-value threshold of 20. FPs were identified as in the false discovery tests described earlier.
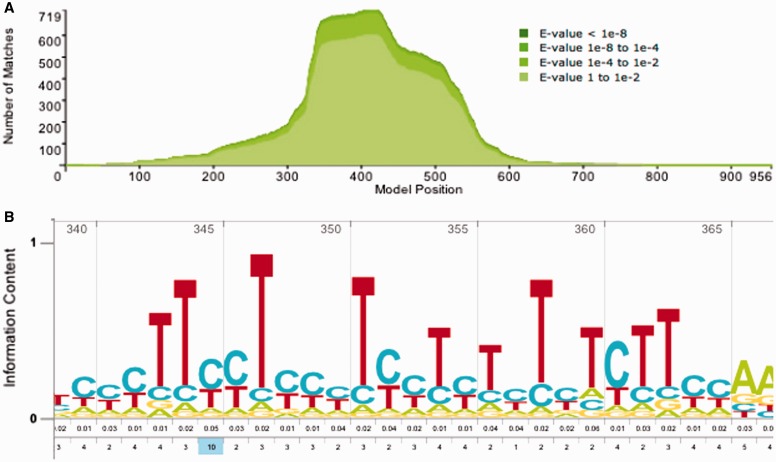
Regions in a model responsible for a large number of FPs were manually inspected, and those with obvious (possibly degenerate) simple repeat patterns were masked in the model. Specifically, in that region of the HMM, emission probabilities were set to match background by (i) masking the corresponding region of the seed using the HMMER3.1 tool alimask then (ii) building a new HMM using hmmbuild. Sequence aligned to the masked region is neither rewarded nor penalized, meaning that only matches supported by bases aligned to flanking sequence will gain sufficient score to be annotated. A true fragmentary instance of a masked model that happens to hit just the masked region will thus not be identified; this is the cost of ensuring non-homologous simple tandem repeats are not incorrectly annotated. A total of 14 models were masked, with one example shown in Figure 3.
Figure 3.
(A) Reverse hit coverage of Tigger16a (DF0000028), before model masking. Without model masking, Tigger16a showed 85 reverse hits with E-value <0.25 (the current FDR-based gathering threshold) against dfamseq-rev, with the highest-scoring reverse hit having an E-value of 2.9e-6. All met the definition of false hits. This is a large fraction of the 3753 Tigger16a hits meeting the same threshold against dfamseq. False hits were focused on one part of the model, between positions 300 and 550. (B) HMM logo of the model region responsible for most of the score of false hits. Model positions 344–361 (shown) exhibit properties of a degenerate simple tandem repeat. By masking this short block of the model, only five false hits more significant than E-value of 0.25 remain, with none more significant than 0.007. Masking caused the loss of 145 (of 3753) dfamseq hits meeting the same score threshold.
Two score thresholds were then established for each model. The ‘gathering’ (GA) threshold is appropriate for annotating the human genome, when knowledge of each TE’s approximate copy number may be used to tune the threshold to yield high sensitivity with low FDR. The ‘trusted cut-off’ (TC) threshold is more stringent, and is intended for use when annotating other organisms, whereas the moderately permissive gathering threshold might lead to a small number of false matches even when no true matches are found. For example, it is not a problem if several dozen of the roughly 1 million human Alu matches are FPs, but finding a few Alu matches in a fish genome would be surprising and misleading. Therefore, the gathering threshold should be used for human searches with the Alu model (higher sensitivity), whereas the trusted cut-off should be used when searching an Alu model against a fish genome. When searching Dfam models with nhmmer, the GA threshold is accessed using the flag ‘–cut_ga’, and the TC threshold is accessed using ‘–cut_tc’.
To establish these thresholds, all models were searched again versus dfamseq and dfamseq-rev, with an E-value cut-off of 20. The GA threshold was established using the methods described in the ‘Sensitivity and False Discovery’ section, using a target empirical FDR of 0.2% and maximum E-value of 20. The TC threshold is at least as high as necessary to reach an E-value of 0.0001 for that model, and is adjusted upwards so that it is always higher than any FP hit on dfamseq-rev (i.e. an empirical FDR of 0).
In some cases, users may be willing to accept much higher rates of false positive annotation. The infrastructure of the Dfam website does not support this, but model libraries with theoretical FDR of 1% and 5% (not E-value constrained) have been made available in the supplementary material (see the ‘Availability’ section). The score thresholds for models in these libraries are stored in the so-called “noise cut-off (NC)” field of the HMM, accessed in nhmmer with the flag ‘–use_nc'. Relaxing thresholds in this way is more sensitive, but also more susceptible to FPs. For example, if hits are allowed with E-values up to 100 with FDR 0.2%, coverage increases just over 8 million bases on the benchmark, but at the cost of more than 1 million false positive bases.
WEBSITE
On the Dfam website (http://dfam.janelia.org) users can browse Dfam entries, jump quickly to an entry via a ‘jump to’ box, find entries via a keyword interface found on all pages, download models and seed alignments of all entries, submit a sequence to the Dfam search interface and access a collection of data views that provide insight into the construction, sensitivity, relationships and distribution characteristics of each entry.
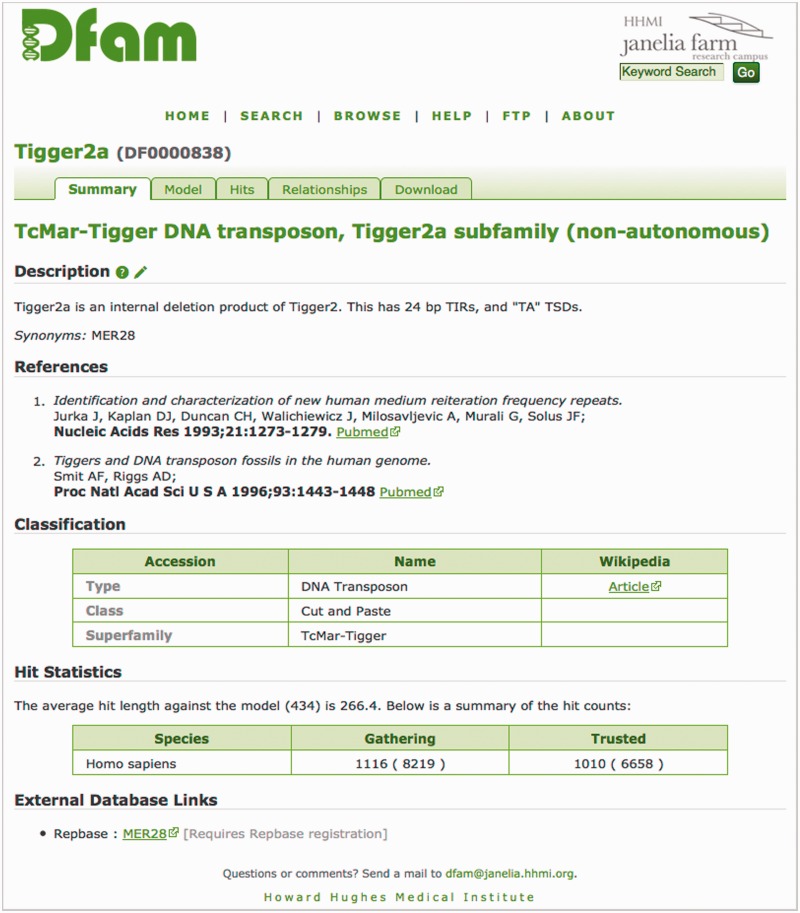
Everything related to a Dfam entry is collected on a single page, which is sub-divided into tabbed panes. Figure 4 shows a typical page for a Dfam entry, with tabs for Summary information, Model data, Hit distribution details, Relationships between models and Downloads for the model, seed and hit lists. The Summary information includes a brief description of the entry, a three-tier classification including links to Wikipedia article when available and number of matches to the model found in dfamseq with scores above the curated ‘gathering’ threshold and above the more stringent ‘trusted cut-off’. Where appropriate, references, links to external database entries and list of synonyms are included. External contributors may add to the annotation of an entry by submitting text via the annotation submission form associated with the description, or indirectly by editing the Wikipedia article that contains more detailed functional annotation.
Figure 4.
A Dfam entry page from the website. This page shows the summary information for Tigger2a (DF0000838). The tabs at the top allow users to browse the different types of associated information.
When viewing an entry, two warnings should be heeded. (i) Some of the Dfam entries represent either tandemly repeated satellite DNA or non-coding RNA genes (ncRNAs) or their pseudogenes, not TEs. The ncRNA entries are included to prevent inappropriate annotation, either because a paralogue has been incorporated as part of a TE, or because their high copy number could be misconstrued as a TE by ab initio methods. An alert is visible next to the name field for ncRNA entries. (ii) Two types of hit counts are presented for each model: non-redundant and redundant. A particular subsequence matching this model may also match other HMMs, resulting in what we call ‘redundant profile hits’ (RPHs). On the summary page, the first (smaller) count represents the number of matching subsequences for which this model is deemed to be the best among all RPHs; the second (larger) count represents the total number of hits to this model, including sequences that are better explained by some other model. For example, in Figure 4, there were 8219 hits to Tigger2a with score above the gathering threshold, but only for 1116 of them was Tigger2a the highest-scoring hit.
Model tab
The Model tab presents a number of analysis tools and resources intended to broaden understanding of the curation process, with the goals of improving existing models and facilitating future curation efforts. The model page presents a number of tools built with these aims in mind:
An HMM logo (such as that shown in Figure 3B) represents the per-position residue and indel conservation of the HMM for that entry. Each position in the model is represented by a stack of letters, with stack height indicating the information content of the position. The rate and expected length of insertions after each position are shown in the fields below each stack. The logo can be zoomed to show more or less of the model as desired.
The consensus sequence derived from the HMM — this will often agree with the original consensus sequence used to produce the HMM, but may differ in the case that the seed alignment supports an alternate majority character at some positions in the alignment.
The Reverse Coverage plot (Figure 3A) shows how matches to reversed genomic sequence (as described in the ‘Sensitivity and False Discovery’ section) are distributed across the model. This plot helps identify model regions that are responsible for generating FP hits, for example, because of low complexity or simple repeat characteristics. When a region with high reverse coverage is clearly a simple repeat, it should be masked as described earlier. Otherwise, these regions may identify positions in the model/seed alignment that require more careful attention in the curation phase, or perhaps just explain why an entry has particularly high FDR-based score thresholds. Not all reverse hits are false positives, as models sometimes match reverse (not-complemented) copies of themselves.
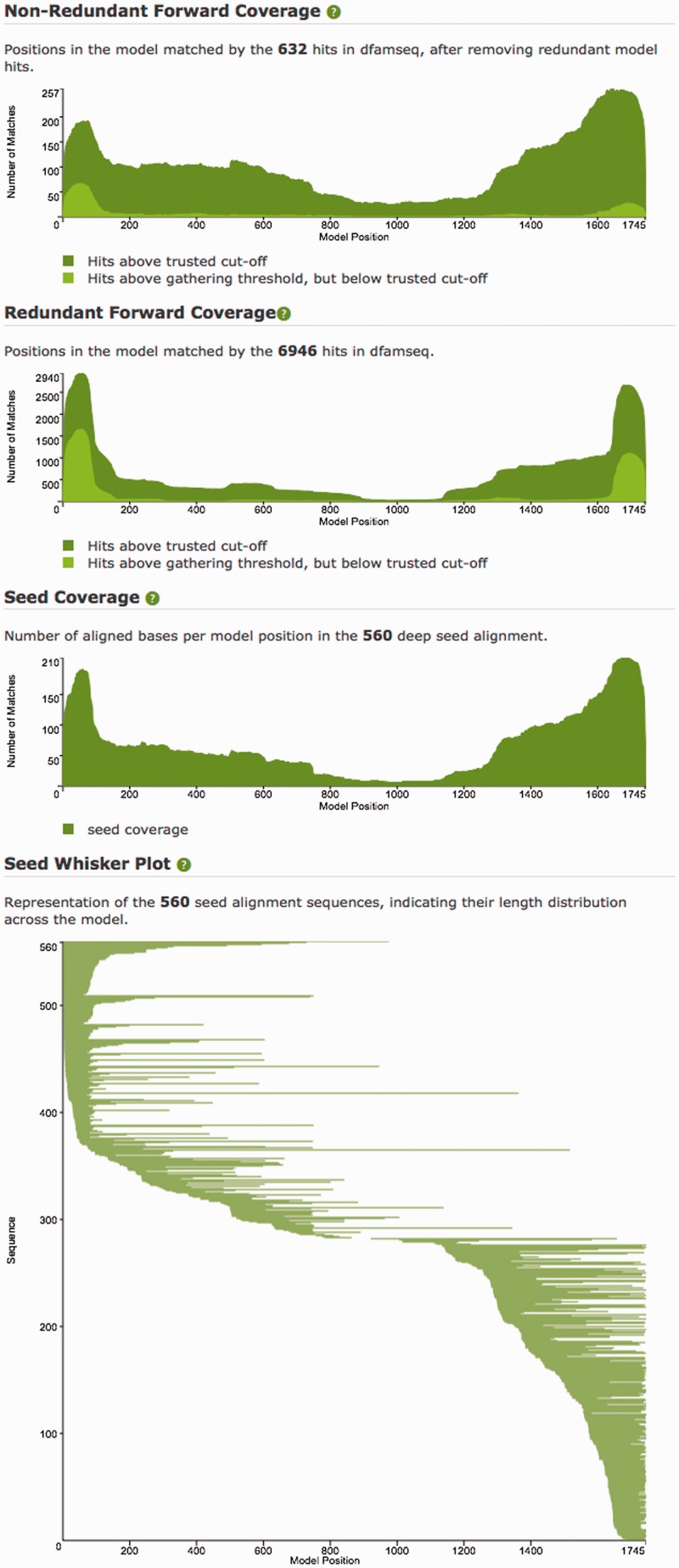
The Forward Coverage plots (Figure 5) show how all above-threshold matches of the model to dfamseq are distributed across the model. Two versions of the plot are given, one showing Non-Redundant Forward Hits (sequences for which this model’s hit is the highest scoring), and the other showing Redundant Forward Hits (all sequences with hit score above the gathering threshold). The Non-Redundant plot often highlights interesting biology, as in the case of Kanga1 shown in Figure 5, in which internal deletions lead to reduced interior coverage. The Redundant plot will often include a large bump in coverage corresponding to a fragment of the TE that shares homology with a related TE (for example, with MIR elements producing many hits to the 3′-end of the L2 model).
The correlated Seed Coverage and Seed Whisker plots (Figure 5) present two perspectives on the way an entry’s seed sequences cover the model’s full length. The Seed plots represent the instances that were used to produce the profile HMM (see the ‘Dfam Entry Curation Process’ section). Thin coverage on a model region, as seen around position 900 of the Whisker plot of Figure 5, can highlight difficulties encountered in seed construction.
Figure 5.
Plots from the Dfam model page for Kanga1 (DF0000218). The Seed Coverage and Whisker plots show that this seed alignment is made of mostly relatively short fragments, and that the middle section of the model is spanned by only a few instances. The Forward Coverage plot shows a common signal for DNA transposons, with the interior portion of the model covered by fewer instances than the termini, as non-autonomous TEs can suffer various degrees of internal deletion, yet must retain critical terminal features. Many of the 5′ terminal hits fall between the gathering threshold E-value of 15 and trusted cut-off E-value of 0.0002, leading to a terminal light green bulge on the left side of the Non-Redundant Forward coverage plot.
Hit tab
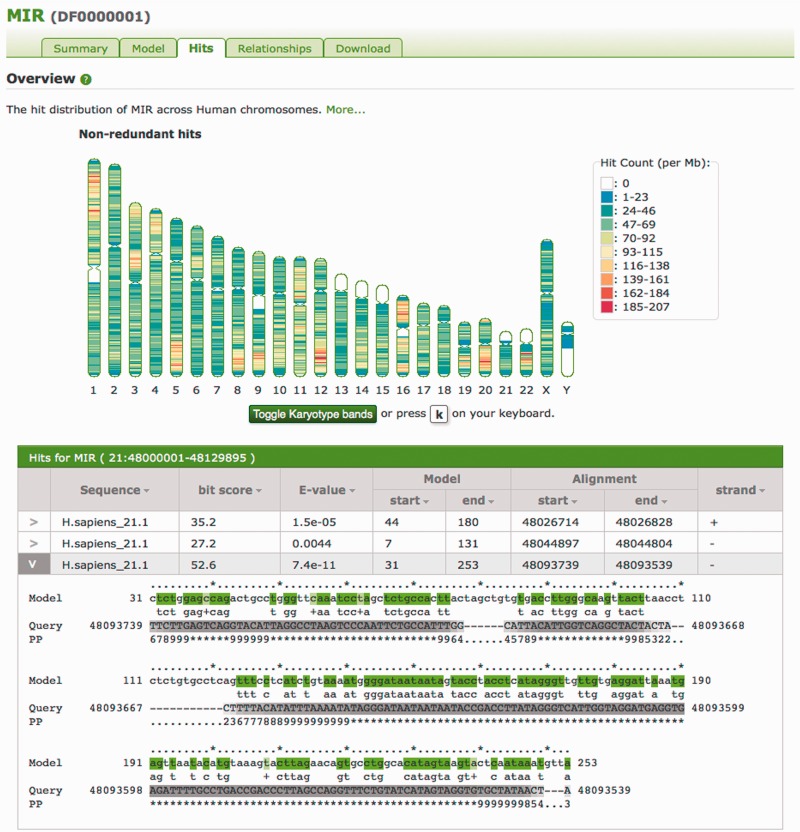
As many TE entries match hundreds of thousands of instances in the human genome, it is difficult to provide all matches via a web interface or as a multiple sequence alignment. To provide access to the matches, a graphical interface has been developed that presents the distribution of hits organized on a karyotype ideogram (Figure 6). Hits are binned in 1-Mb regions, with counts in each bin distinguished by colour. When a region of the hit distribution ideogram is clicked, the hits in that region will be loaded below the karyotype ideogram. Each hit can be expanded to reveal the alignment between the hit sequence and the model. A full listing of hits (but not alignments) for an entry may be retrieved from the download page. By default, the page represents non-redundant hits, but the image and hit lists can be toggled to show redundant hit distributions. The ideogram can also be toggled to reveal the canonical Giemsa stain banding of the chromosomes, providing a reference to positional context. It is important to remember TEs split by more recently mobile TEs have not been aggregated as would be the case with RepeatMasker’s expert system post-processing, so older fragmented TEs may be counted as multiple instances.
Figure 6.
The Hits tab for the MIR (DF0000001) entry. This graphic shows the non-uniform distribution of MIRs across the human genome. Large patches of white in the hit distribution ideogram indicate regions with no instances of the model; in this case, these are particularly difficult to sequence heterochromatic regions (represented by N’s in the genome sequence) as can be seen by toggling karyotype bands. Below the karyotype ideogram are given the hits from a region on chromosome 21, with one hit expanded to show the alignment of that hit to the MIR model. In the alignment, the model line presents the consensus sequence for aligned states in the model, coloured according to the match line. The PP line represents the posterior probability, or degree of confidence in each aligned residue (for example, with ‘*’ meaning highest confidence, and low numbers indicating low confidence), with corresponding grey scale colouration of the Query sequence.
Relationships tab
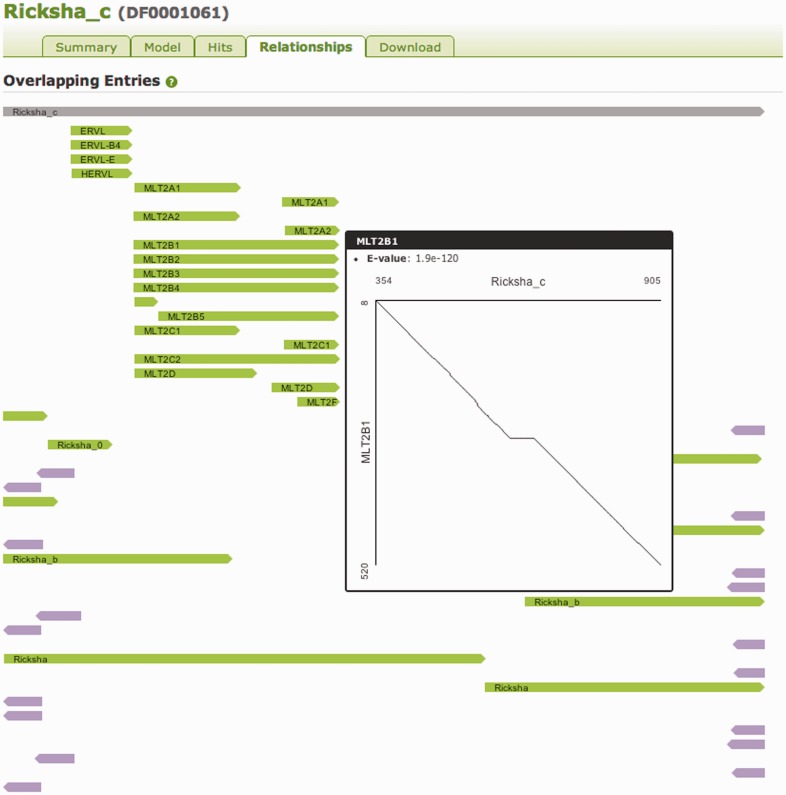
Many models have a complicated relationship with other models, as in the cases of Ricksha (which long ago picked up the 3' end of an ERVL, including its LTR, MLTB2), and SVA (which carries copies of both a portion of a HERVK LTR and two Alus in reverse orientation). The Relationships tab presents a graphical aid to understanding such cases of hitchhiking as well as the relationships between autonomous and non-autonomous elements (such as the MER104 and Kanga1), and the more straightforward relationships between subfamily entries. An example of a Relationship tab is shown in Figure 7.
Figure 7.
The Relationship tab for the Ricksha_c (DF0001061) entry. Consensus sequences were produced for all models using the HMMER3 tool hmmemit. These sequences were then searched with all models using nhmmer, with a hit with E-value better than 1e-5 supporting a relationship. Simple glyphs are used to represent the location of different TEs along the along the model, indicating orientation by shape and colour. In this case, the relationships to the ERVL and MLT2 subcomponent elements are represented, as are relationships to other Ricksha models. Placing the mouse over one such glyph raises a dot plot (12) that shows how these elements align to each other.
Search page
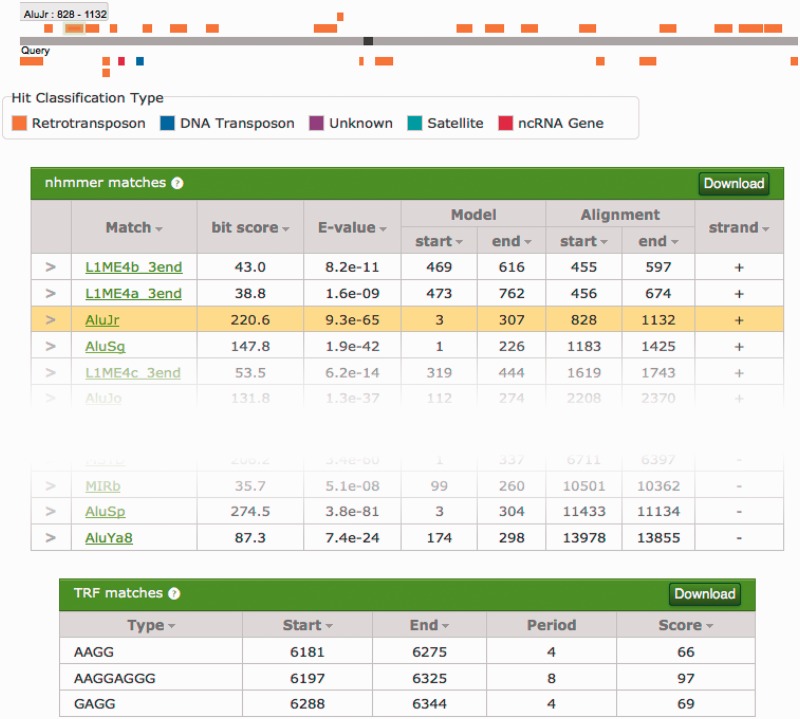
A user may submit a sequence of length up to 50 kb to the Dfam search interface, accessed via the menu at the top of every page. The search consists of two parallel phases: tandem repeat identification with TRF and a scan with all Dfam models. In the case of RPHs, in which multiple Dfam models hit the same portion of the submitted sequence, shorter and lower-scoring hits are ignored. These conservatively non-redundant Dfam hits, along with TRF matches, are presented in both tabular and graphical representation, as shown in Figure 8. The script used to resolve RPHs, called dfamscan.pl, is available for download via the FTP site.
Figure 8.
Example of a user-submitted search result. The submitted sequence is represented by the top grey bar, with overlaid black boxes representing TRF matches. Non-redundant Dfam hits to the plus strand are organized above the sequence bar, and hits to the minus strand are organized below the bar. The colour of each Dfam bar depends on the entry type (DNA transposon, RNA retrotransposons, ncRNA, etc.). When a bar is clicked, the row corresponding to that hit is highlighted on the page.
The Retrieve Hits tab enables visualization of the pre-calculated hits to ≤50-kb regions of dfamseq (i.e. to select regions of human chromosomes). RPHs can be resolved using the same procedure as used by dfamscan.pl, as desired. This search can also be restricted to a single Dfam entry, with or without RPH resolution.
GENOME ANNOTATION
In the first release of Dfam, we aimed to highlight features of the new database, to enable assessment of the impact of profile HMMs on TE search sensitivity and to allow downstream usage of the database for full annotation of an entire genome (human) by RepeatMasker. The HMM database may be downloaded from the FTP site (ftp://selab.janelia.org/pub/dfam/Current_Release). Supporting software is also available for download, including nhmmer and dfamscan.pl. The dfamscan.pl script also forms the basis of the web search, but should be treated as a first pass at annotation, not as a replacement for RepeatMasker, which is a more thorough expert system that incorporates Dfam and nhmmer.
When searching with Dfam models, it is important to remember that the gathering threshold (accessed using the nhmmer flag ‘–cut_ga’) is appropriate for annotating the human genome, and the trusted cut-off (‘–cut_tc’) is appropriate for non-human genomes. See the ‘Model Masking and Thresholds’ section for details.
Defining coverage as was done for the results in Table 1, searching with Dfam and nhmmer produces 54.48% coverage of unambiguous chromosomal sequence in human (1 559 503 431 bases). This number will be improved through improvement to existing models, addition of new models representing as yet unidentified TE families, judicious changes to gathering thresholds and application of expert system downstream analysis (such as in RepeatMasker, which, for example, uses the approach of clipping young elements out of genomic sequence before searching for older elements).
Recent results combining a method called P-clouds (13) with RepeatMasker results suggest that at least 66% of the human genome is derived from ancient TE copies. We believe this finding is noteworthy, but observe that for the purposes of that study, the authors were willing to tolerate a ∼17% FDR to maximize detection sensitivity, with >40% of P-cloud-specific predictions estimated to be FP. For our goal of reliable annotation of parts of the genome that are derived from known TE families, our benchmark and evaluation are performed at much greater stringency, so our coverage statistics are not directly comparable with those in (13).
CONCLUSION
Dfam and nhmmer have been incorporated into RepeatMasker, for use in annotating human genomes. We are currently developing a BigBED file (for use at UCSC) and DAS server (for use at Ensembl), as well as a new UCSC RepeatMasker track, to enable visualization of Dfam data in genome browser context.
Repbase contains consensus sequences for TEs from dozens of organisms, and will continue to be an invaluable resource for new entries and annotation updates. The protocol we used to produce the 1143 current Dfam entries can be expanded to the remainder of Repbase consensus sequences, allowing TEs from a broad range of organisms to be added to Dfam. While expanding species breadth, we will test the expected benefits of incorporating seeds built from slower-evolving organisms.
In addition to broadening the scope of Dfam to include more species, we will aim to reorganize the handling of redundant models, to better leverage the ability of profile HMMs to represent families of sequences. We will also shift the accumulated knowledge of RepeatMasker’s model name mapping out of software and into the database.
The curation toolkit has currently been used only by the Dfam curators, but has been constructed with community curation in mind, as is possible with Pfam. In the near future, our infrastructure will enable significant external contribution to Dfam by placing our collection of curator-assistance tools in the hands of the community of TE experts. As the Dfam library grows, model consensus sequences and annotation will be fed back to Repbase to synchronize these two important TE annotation resources.
AVAILABILITY
The Dfam website site is available at http://dfam.janelia.org. Dfam data can be freely downloaded from the FTP site (ftp://selab.janelia.org/pub/dfam/) either as flat files or in the form of MySQL table dumps. The software nhmmer is available via the FTP link at the top of every Dfam web page. Data and scripts used to produce Tables 1 and 2, as well as per-family coverage results and high-FDR model libraries, can be downloaded at http://selab.janelia.org/publications/Wheeler13/Supplementary_material.tar.gz.
FUNDING
Howard Hughes Medical Institute Janelia Farm Research Campus (to R.D.F., J.C., S.R.E., T.A.J. and T.J.W.); National Institutes of Health [P41LM006252-1 to J.J., RO1 HG002939 to A.F.A.S. and R.H.]. Funding for open access charge: HHMI Janelia Farm Research Campus.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the anonymous reviewers for their insightful and constructive suggestions. Goran Ceric provided masterful support of Janelia Farm’s high-performance computing resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
REFERENCES
- 1.Jurka J, Klonowski P, Dagman V, Pelton P. CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem. 1996;20:119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- 2.Smit AFA. 1995. Structure and evolution of mammalian interspersed repeats. Ph.D. Thesis. University of Southern California. [Google Scholar]
- 3.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogent. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 4.Park J, Karplus K, Barrett C, Hughey R, Haussler D, Hubbard T, Chothia C. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- 5.Durbin R, Eddy SR, Krogh A, Mitchison GJ. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 6.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, McWilliam H, Goujon M, Cowley A, Lopez R, Pearson WR. PSI-Search: iterative HOE-reduced profile SSEARCH searching. Bioinformatics. 2012;28:1650–1651. doi: 10.1093/bioinformatics/bts240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs AJ, McIntyre GA. The diagram, a method for comparing sequences. Its use with amino acid and nucleotide sequences. Eur. J. Biochem. 1970;16:1–11. doi: 10.1111/j.1432-1033.1970.tb01046.x. [DOI] [PubMed] [Google Scholar]
- 13.de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]