Abstract
The BRENDA (BRaunschweig ENzyme DAtabase) enzyme portal (http://www.brenda-enzymes.org) is the main information system of functional biochemical and molecular enzyme data and provides access to seven interconnected databases. BRENDA contains 2.7 million manually annotated data on enzyme occurrence, function, kinetics and molecular properties. Each entry is connected to a reference and the source organism. Enzyme ligands are stored with their structures and can be accessed via their names, synonyms or via a structure search. FRENDA (Full Reference ENzyme DAta) and AMENDA (Automatic Mining of ENzyme DAta) are based on text mining methods and represent a complete survey of PubMed abstracts with information on enzymes in different organisms, tissues or organelles. The supplemental database DRENDA provides more than 910 000 new EC number–disease relations in more than 510 000 references from automatic search and a classification of enzyme–disease-related information. KENDA (Kinetic ENzyme DAta), a new amendment extracts and displays kinetic values from PubMed abstracts. The integration of the EnzymeDetector offers an automatic comparison, evaluation and prediction of enzyme function annotations for prokaryotic genomes. The biochemical reaction database BKM-react contains non-redundant enzyme-catalysed and spontaneous reactions and was developed to facilitate and accelerate the construction of biochemical models.
INTRODUCTION
BRENDA (BRaunschweig ENzyme DAtabase, http://www.brenda-enzymes.org) is the major information system for enzyme-related research. The development was started 25 years ago and the data were made available via the internet in 1998 with a first query system. Since then it has been continually updated and further developed to meet the requirement in newly arising branches of biomedical research like systems biology or metabolomic research. The database holds a wide range of aspects of enzymology such as functional data like enzyme-catalysed reactions, kinetic data for catalysis and enzyme inhibition, enzyme stability, purification, crystallization or mutations, as well as the largest collection of enzyme names and synonyms, altogether stored in ∼50 information fields. Each data entry is connected to the literature reference, to the name of the source organism, and to the protein sequence identifier (if available). For many organisms the cell type or a strain specification is included. Enzymes from multicellular organisms (e.g. mammals or plants) are further specified with respect to their occurrence in body parts, organs or plant anatomy. The terms used here are based on the BRENDA Tissue Ontology (BTO) (1). This reference system has been developed in parallel to the enzyme database as an encyclopedia for tissue terms and cell types including synonyms and definitions and currently holds ∼5300 different terms. The subcelluar localization of an enzyme is described using the terms of the Gene Ontology (GO) (2).
The term ‘ligand’ is used in BRENDA for all compounds, which interact with enzymes. These can be small molecules like the metabolites in the primary metabolism, macromolecules, cosubstrates/cofactors or metal ions which must be present for the activity of the enzyme. Enzyme inhibitors represent a major portion of the ligands. These can be of variable origin like naturally occurring antibiotics or synthetic chemicals analysed for the development of drugs or pesticides. Macromolecules like DNA, RNA, proteins or polysaccharides also interact with enzymes as substrates, inhibitors or as agents that regulate and modify the activity. The affinity of individual ligands for an enzyme is stored in BRENDA in different kinetic or association constants as KM, kcat, KM/kcat for substrates or Ki, IC50 values for inhibitors.
For more than 130 000 BRENDA ligands the chemical structure is stored. This allows the display of structural diagrams for the molecules and the chemical reactions. All stored ligand structures are searchable via a query based on a structure or substructure entered by the user. This is essential because of the variety of different names being used for compounds. On average ∼20% of the ligands in BRENDA are stored with different names, some with 20 different names like the common adenosine 5′-phosphate (AMP). Using the structural information the ligands in BRENDA are then linked to the ChEBI (3) database of chemical entities of biological interest.
In many cases the biochemical reaction is defined using generic terms for molecules. Examples are (1->4)-α-D-glucooligosaccharide or aryl-β-D-glucoside. They are stored in the database using the Markush concept (with R as generic substituents). Since the huge amount of publications on enzyme properties does not allow the manual annotation of the complete literature of all enzymes, the manually annotated data are supplemented with information retrieved by text mining procedures. Three supplementary databases therefore complete the information. The quality level of automatic retrieved information still remains behind manually annoted data. However text mining techniques are improving. Thus the deployed text mining procedures for the automatic generated data bases are continually adapted to new findings. Additionally entries of these data bases are accompanied by reliability categories and quality scores. These enables the user to judge intuitively on the validity of the provided information. The procedures are based on the text interpretation of sentences containing enzyme names, organism names, localization, and source tissues in abstracts and titles of the PubMed database (4). Although the use of the common name is strongly recommended by the IUBMB (International Union of Biochemistry and Molecular Biology) biochemical nomenclature committee, by now we found that more than 65 000 different names are in use for the enzymes of ∼4800 approved EC classes (EC numbers issued by the IUBMB) (5,6). These names were collected via the manual annotation of ∼120 000 literature references and are stored as synonyms. This insures that an enzyme is even found in the database if a rarely used name is entered as a query. For some EC classes such as the protein tyrosine kinase (EC 2.7.10.1) more than 1300 names are found in the literature. On average each EC class has 14 synonyms. For the application in the text mining process this list is curated in order to remove any non-specific enzyme names. Other controlled vocabularies used in BRENDA are the BTO for tissue distribution, the GO for subcellular localization and the NCBI Taxonomy tree (7).
FRENDA (Full Reference ENzyme DAta) aims at providing an exhaustive collection of indexed literature references containing organism-specific enzyme information by providing all combinations of enzyme names and organisms found in PubMed titles or abstracts together with the literature citation (8).
AMENDA (Automatic Mining of ENzyme DAta) is a subset of FRENDA and comprises enzyme-specific information on the enzyme source based on the vocabulary of the BTO and the subcellular localization based on the GO terms. The results are classified into four reliability categories depending on the occurrence of search terms in title and/or abstract and/or MeSH terms.
KENDA (Kinetic ENzyme DAta) was developed to include additional functional kinetic enzyme data. This text mining approach extracts kinetic values and expressions from more than 2.2 million PubMed abstracts based on the results of the FRENDA database.
DRENDA provides broad information on the connection of diseases and enzymes. It is based on the analysis of disease-related enzyme information using a subset of MeSH terms. The results are classified into the categories causal interaction, therapeutic application, diagnostic usage, and ongoing research and presented together with the respective quality scores (9).
For a complete picture on the enzyme properties BRENDA includes data from many external sources such as genome sequences [EBI Genomes Server (10), Ensembl database (11)], protein sequences [UniProt databases (12,13)], functional assignments [COG database (14)], molecular pathways [KEGG database (15)] and protein 3D-structures from the PDB (16). Links are provided for individual enzymes to metabolic databases such as KEGG or MetaCyc (17) and to the enzyme nomenclature of the IUBMB. Computer programs are used for the prediction of membrane-associated enzymes, for the display of active centers or the cofactor binding sites, or for the display of the enzymes in the genomic context (Genome Explorer).
In this article, we give an overview on the enzyme data in BRENDA, AMENDA, FRENDA, and the newly developed databases KENDA and DRENDA. Likewise available from the website are the newly developed integrated biochemical reaction database BKM-react (18) and the Enzyme Detector (19) which are described in detail.
BRENDA DATA
Enzyme classification
The database covers all enzyme classes that have been classified by the IUBMB plus more than 300 others that are not yet characterized well enough to be fully classified. Currently there are 4867 active EC classes plus 871 EC classes which have been deleted or transferred to other EC classes due to new research results. The number of EC classes is rising constantly.
In the course of the manual literature annotation process for BRENDA we frequently find enzymes that are not yet classified. These are enzymes that differ substantially in substrate specificity and reaction from all current EC classes. They are enzymes closing a gap in a metabolic pathway or enzymes of a new pathway detected in a specific organism. These enzymes have to be classified within the hierarchical EC-system and therefore are prepared within the BRENDA team for the final decision of the IUBMB Enzyme Commission with two active members of the BRENDA team. Even before classification the newly detected enzymes are integrated into the BRENDA database as ‘preliminary BRENDA-supplied EC number’. Instead of the common EC number these entries carry a B and a serial number in the fourth position of the EC number. Currently (July 2012) BRENDA holds 321 preliminary EC numbers. A portion of these numbers is under review at the IUBMB. All preliminary BRENDA EC numbers are internally reviewed regularly and where available new data are added. Table 1 shows the preliminary BRENDA-supplied entries of aspartic proteinases. None of these are currently under review by the IUBMB because there is an ongoing discussion on how to classify proteinases and how to describe their specificity.
Table 1.
Preliminary BRENDA-supplied entries of aspartic proteinases
| EC-class | Name | Reaction and specificity |
|---|---|---|
| 3.4.23.B6 | Mason-Pfizer monkey virus proteinase | The enzyme cleaves 17 amino acids of the C-terminal 38-amino-acid cytoplasmic tail of the transmembrane protein TM of the released immature virus. |
| 3.4.23.B10 | Rous sarcoma virus retropepsin | The cleavage sequence in the natural substrate NC-PR is PPAVS-/-LAMTMRR. The activity can be improved by substitution by Trp, Tyr, Phe, Leu, Arg, Glu, His or Ala in P1, Tyr in P3′, and Arg, Phe, Asn or His in P3. |
| 3.4.23.B11 | Spumapepsin | Good cleavage at the peptide bonds: Asn-Thr, Asn-Gln, Asn-Cys and Asn-Ala. |
| 3.4.23.B13 | Proteinase P15 | Efficient cleavage of Ala-Thr-His-Glu-Val-Tyr-Phe(NO2)-Val-Arg-Lys-Ala, no cleavage with Ser, Arg or Glu at P1, Gly or Phe at P2, and Pro at P3. Specifically liberates the five major structural proteins from the common gag precursor, as well as reverse transcriptase and integrase from the gag-pol precursor. |
| 3.4.23.B14 | Plasmepsin IV | Cleavage of hemoglobin. In the S3 and S2 subsites, the plasmepsin 4 orthologs all prefer hydrophobic amino acid residues, Phe or Ile, but reject charged residues such as Lys or Asp. In S2′ and S3′ subsites these plasmepsins tolerate both hydrophobic and hydrophilic residues. |
| 3.4.23.B2 | Simian immunodeficiency virus proteinase | The enzyme may have a wide substrate specificity. Good cleavage of the peptide bonds Met-Met and Tyr-Pro. Cleavage is also observed at Phe-Pro, Phe-Leu, Leu-Phe, Leu-Ala, Glu-Ala and Tyr-Ala. |
| 3.4.23.B3 | Equine infectious anemia virus proteinase | Processing at the authentic HIV-1 PR recognition site and release of the mature p17 matrix and the p24 capsid protein, as a result of the cleavage of the -SQNY-/-PIVQ- cleavage site. |
| 3.4.23.B4 | Feline immunodeficiency virus protease | The enzyme seems to have a preference for Val in P1′ and Phe in P1. In contrast to the HIV-1 protease the feline immunodeficiency virus protease does not cleave the peptide KSGVFVQNGLVK at the Phe-Val bond. Gln in P2′ may be inhibitory. In contrast to HIV-1 protease the feline immunodeficiency virus protease does not cleave peptide KSGNFVVNGLVK at the Phe-Val bond. Asn in P2 may be inhibitory. |
| 3.4.23.B5 | Murine leukemia virus protease | Processing of viral polyprotein. The retroviral protease is essential for virus replication, by processing of viral Gag and Gag-Pol polyproteins. |
| 3.4.23.B8 | Human T-cell leukemia virus type 1 protease | Processing at the authentic HIV-1 PR recognition site and release of the mature p17 matrix and the p24 capsid protein, as a result of the cleavage of the -SQNY-/-PIVQ- cleavage site. |
| 3.4.23.B9 | Bovine leukemia virus protease | The best substrate YDPPAILPII is bearing the natural cleavage site between the matrix and the capsid proteins of BLV Gag precursor. polyprotein. Good cleavage of the peptide bonds: Leu-Pro, Leu-Val, Gly-Val and Leu-Pro. |
| 3.4.23.B1 | Napsin | proteolytic cleavage of polypeptides to large and stable peptides. |
| 3.4.23.B17 | Walleye dermal sarcoma virus proteinase | Processing of viral polyprotein. Preference order for P1 position is Phe > Tyr > Leu, Met > Ala. Gly is preferred at position P3. Ala and Pro are preferred at position P4. Asn, Cys or Leu are preferred at position P2. |
| 3.4.23.B18 | Mouse mammary tumor virus retropepsin | Processing of viral polyprotein. Selective for large aromatic residues (Tyr and Phe) at position P1. Phe and Leu are preferred at position P3. No hydrolysis of substrates with Gly or Ala at position P3. Medium-sized or large hydrophobic residues as Ile, Leu and Phe are preferred at position P4. |
| 3.4.23.B19 | Plasmepsin V | Cleavage of hemoglobin. In contrast to the food vacuole plasmepsins, detergent-solubilized PMV does not bind the aspartic protease inhibitor pepstatin. |
| 3.4.23.B20 | HycD peptidase | This enzyme specifically removes a 15-amino acid peptide from the C-terminus of the precursor of the large subunit of hydrogenase 2 [UniProt: P0ACE0] in E. coli. |
BRENDA content
The BRENDA information system covers a wide range of enzyme data for each EC class. In this respect it is different from other enzyme databases which are specialized either on the nomenclature [ExplorEnz, ENZYME (20), IntEnz (21)], on the metabolic pathways (e.g. KEGG, MetaCyc) or on specific classes of enzymes [e.g. Merops (22), MODOMICS (23), CAZy (24), Kinomer (25)].
The data are either manually extracted from the primary literature, or obtained from data integration of data from other sources or obtained by text mining (and disclosed as such). Each single entry is connected to a reference, covering the literature between 1939 and 2012. It is also connected to an organism, and where available to a strain and a sequence identifier for the enzyme-protein. This is stored in a database with more than 200 million data entries, 2.7 million of which are hand-annotated and stored in ∼50 categories. These are grouped into ‘Nomenclature’, ‘Reaction and Specificity’, ‘Isolation and Preparation’, ‘Functional Parameters’, ‘Organism-related Information’, ‘Stability’, ‘Enzyme Structure’, ‘References’ and ‘Application & Engineering’. Each EC class is updated annually, selected new references are annotated and included. Table 2 shows the increase of the amount of data since 2008 in selected areas of the database.
Table 2.
Increase of the manually annotated data content in BRENDA from 2008 to 2012
| 2008 | 2012 | Increase from 2008 to 2012 (%) | |
|---|---|---|---|
| EC classes | 4824 | 5372 | 11.4 |
| Enzyme names | 59 341 | 85 399 | 43.9 |
| Organisms | 8930 | 10 511 | 17.7 |
| Source/tissue | 53 547 | 87 632 | 63.7 |
| Localization | 21 857 | 30 894 | 41.3 |
| KM-values | 89 012 | 121 298 | 36.3 |
| Ki-values | 19 018 | 33 819 | 77.8 |
| kcat-values | 29 149 | 49 224 | 68.9 |
| Mutant properties | 30 437 | 59 841 | 96.6 |
| Ligand names | 119 315 | 177 850 | 49.1 |
| Ligand structures | 48 651 | 103 222 | 112.2 |
| Substrates/products | 244 236 | 324 591 | 32.9 |
| Inhibitors | 127 146 | 203 727 | 60.2 |
| Stability information | 36 683 | 44 716 | 21.9 |
| References | 84 607 | 126 405 | 49.4 |
The numbers refer to the combination of enzyme-organism-(protein)-value. The numbers of the Ligand Names and Ligand Structures, the Organism and the References specify the unique entries in BRENDA.
The section ‘Reaction and Specificity’ lists all reactions which have been found in the annotated literature. These reactions are not restricted to naturally occurring substrates but also comprise synthetic substrates. The latter give valuable information on possible applications of the enzyme in biotechnology, in the food industry or for agricultural purposes. Synthetic substrates are also widely used for the determination of substrate specificity. The reactions, substrates, products, inhibitors or cofactors can be displayed as chemical diagrams.
Kinetic data are stored to evaluate the efficiency of an enzyme and provide insights into the catalytic process. In BRENDA several kinds of kinetic data are stored: KM, kcat, Ki, IC50 values. In addition to the numerical value, recalculated to standard units, and the substrates or inhibitors these data fields contain essential information stored in the commentary section, as e.g. information on the experimental conditions, the isoenzymes or mutant forms etc. The comparison of kinetics for wild-type, mutant or enzymes produced by site-directed mutagenesis gives insight into the catalytic process. When a mutation is associated with a disease the altered kinetic behavior can lead to valuable conclusions and possibly to a treatment. In BRENDA such data can be accessed for example in the KM search field by typing ‘mutant’ in the commentary search box. The optimal temperature and pH are also given. However the kinetic data are not always recorded at the optimal temperature and pH, for experimental reasons such as the instability or insolubility of the substrates.
Organism-related data
Each data entry in BRENDA is linked to the name of the source organism and, where available to a strain name and a protein identifier (generally UniProt accession codes). Currently BRENDA stores enzyme data for 10 480 different organisms.
Sequence and structure data
The section ‘Enzyme Structure’ contains 2.37 million links to the protein sequences of the UniProt database and 78 000 3D-structures of the Protein Data Bank. These data are used for the visualization of the 3D-structures using JMOL (26), which is integrated into the BRENDA website. Structural and functional features, such as active sites or binding sites can be displayed in the representation of the enzyme (27).
Sequence data are included into the calculation of the transmembrane helices to provide the prediction of the number, the size and the location of these helices using TMHMM [TransMembrane Hidden Markov Model, (28)].
AMENDA and FRENDA
While the manually annotated BRENDA database aims at selectively providing enzyme functional data it cannot provide a full record and annotation of the complete literature being published in enzymology. The databases FRENDA and AMENDA complement the information with data derived from text mining. FRENDA provides links to all PubMed references that cover enzyme-specific information in combination with the name of the organism or one of its synonyms. AMENDA is a subset of FRENDA and specifies the occurrence in tissues, organs and the subcellular localization. Here the text mining procedure is more refined and the vocabularies have been intensively curated. The results are ranked to four reliability categories. The current FRENDA database stores ∼1.9 million reference data for enzymes. The subset AMENDA contains 1.1 million ranked entries. Of these ∼230 000 give information on the occurence in tissues linked to the BTO and 52 000 define the subcellular localization.
NEW DEVELOPMENTS
Kinetic ENzyme DAta
BRENDA contains more than 285 000 manually annotated kinetic values. Nevertheless they cannot cover the entire enzyme literature. Recently this gap could be filled with data from a text mining method adapted from a previously developed method (29). The procedure is based on text interpretation and supported by dictionaries with ∼2000 collected kinetic terms and units (including different spellings), ∼2000 terms for the interpretation of the sentence structure (e.g. negations, marker for listings). Approximately 180 000 names for metabolites, inhibitors and other compounds are taken from the BRENDA database. A total of 63 103 enzyme names and 5048 EC classes together with 775 684 organisms names and their synonyms from the FRENDA database are included in the search. The method is performed on 2.2 million PubMed abstracts, representing only those where enzymes have been found by FRENDA, and extracts kinetic values in 14 categories (KM, Ki, kcat, kcat/KM, Vmax, IC50, S0.5, Kd, Ka, t1/2, pI, nH, specific activity, Vmax/KM). The procedure involves the following steps:
-
–
Abstracts are split into sentences. Sentences and titles (handled as sentences) are stored in the results database.
-
–
If an organism or an enzyme was found by FRENDA the sentence ID is stored along with the organism and/or enzyme (systematic name or EC class) in the results database.
-
–
Kinetic expressions are extracted from sentences with enzymes.
-
–
Kinetic units are extracted from sentences with kinetic expressions.
-
–
Values, ligands and other terms (binding words, markers for listings etc.) are extracted from sentences with kinetic units.
-
–
Sentences with enzyme, kinetic expression, kinetic unit and number are further processed:
-
–
Nested terms are removed.
-
–
Values and units are merged.
-
–
Values and terms are removed if headed or followed by a marker for removal.
-
–
Lists are identified.
-
–
Remaining sentences are stored into the results database.
-
–
If no organism is found in the sentence and only one organism has been found in the abstract, this organism is handled as if found in the sentence.
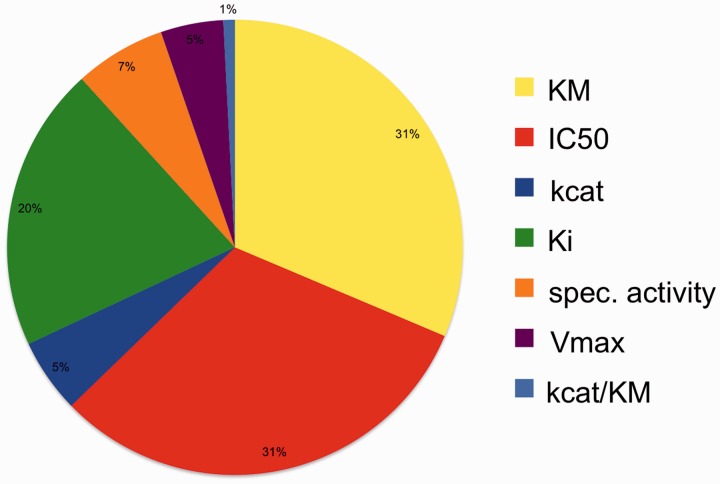
The database contains 20 817 kinetic data stored as kinetic value connected to a kinetic expression, to the unit, to the EC class, to a reference and to a source organism if mentioned in the text. Access to the new data is provided via the homepage and selecting ‘Kinetic ENzyme DAta’. KM and IC50 values represent the large majority of the found values. Figure 1 shows the relative distribution of the kinetic categories in the results. The analysed PubMed abstracts can be displayed to view the context. Figure 2 shows a typical abstract which has been analysed with KENDA.
Figure 1.
Relative distribution of the kinetic categories in the results of the KENDA data.
Figure 2.
Abstract from Pubmed (ID 19090111) for α-glucosidase showing the kinetic values highlighted by the KENDA procedure.
Disease-Related ENzyme information DAtabase
Due to their determinative and crucial role in all aspects of life, including metabolism, regulation, immunity, etc., the absence or malfunction of enzymes leads to severe pathologic conditions in an organism and may manifest itself in a disease.
The Disease RElated ENzyme information DAtabase DRENDA represents a supplemental database to BRENDA including classified literature links and an analysis of enzyme/disease relations. In a first step enzyme-related information on diseases in abstracts of the PubMed database was retrieved by automatic text analysis supported by vocabularies from BRENDA (EC numbers, enzyme names, ∼100 000 items) and MeSH terms from the NCBI for diseases and metabolic disorders (∼22 000 terms). This step resulted in 0.9 million incidences of enzymes and diseases in the literature. Table 3 summarizes results for the most frequently described enzyme–disease relationships and the EC classes which have been found to be connected with the highest number of different diseases.
Table 3.
Enzyme–disease-related data in the DRENDA database
| Disease | PubMed IDs | EC no. |
|---|---|---|
| Neoplasm | 110 485 | 1272 |
| Infection | 25 937 | 1067 |
| Carcinoma | 22 141 | 829 |
| Breast neoplasm | 18 053 | 667 |
| EC class | PubMed IDs | Disease terms |
|---|---|---|
| 2.7.10.1 | 28 683 | 1178 |
| 1.7.2.1 | 13 946 | 1058 |
| 1.14.99.1 | 10 902 | 897 |
| 3.4.15.1 | 11 650 | 689 |
| 3.4.21.68 | 11 939 | 864 |
However, a simple listing of references with enzymes names which are connected with a disease name is not of much value for the researcher. In order to assess the kind of connection or dependency the relationship was classified into four categories:
‘Causal Interaction’ of a disease caused by a malfunction of the enzyme;
‘Ongoing Research’ when the disease–enzyme relationship is suspected but research is still under way;
‘Diagnostic Usage’ when the enzyme is part of the diagnostic course of action like the measurement of its activity, the test for its presence or the assay of its functional characteristic parameters; and
‘Therapeutic Application’ when the enzyme is applied as a therapeutic agent or considered as drug target.
The results of the procedure were evaluated with respect to precision, recall, accuracy and specificity in a 5-fold cross- validation process. This ensures high-quality data and represents a true upgrade of the BRENDA data. Table 4 displays an overview of the data content and the results of the evaluation procedure.
Table 4.
Results of the DRENDA validation process for the classification of the enzyme-disease relationships
| Category | Precision | Recall | F1 Score | Accuracy | Specificity | Entries |
|---|---|---|---|---|---|---|
| therapeutic application 4 | 0.972 | 0.530 | 0.686 | 0.750 | 0.984 | 114 477 |
| therapeutic application 3 | 0.909 | 0.606 | 0.727 | 0.766 | 0.936 | 173 134 |
| therapeutic application 2 | 0.900 | 0.818 | 0.857 | 0.859 | 0.903 | 330 335 |
| therapeutic application 1 | 0.868 | 0.894 | 0.881 | 0.875 | 0.855 | 415 923 |
| ongoing research 4 | 0.800 | 0.229 | 0.356 | 0.571 | 0.939 | 139 238 |
| ongoing research 3 | 0.765 | 0.371 | 0.500 | 0.616 | 0.878 | 252 136 |
| ongoing research 2 | 0.750 | 0.543 | 0.630 | 0.670 | 0.806 | 341 592 |
| ongoing research 1 | 0.720 | 0.686 | 0.702 | 0.700 | 0.714 | 440 043 |
| diagnostic usage 4 | 0.892 | 0.388 | 0.541 | 0.680 | 0.956 | 175 150 |
| diagnostic usage 3 | 0.848 | 0.588 | 0.694 | 0.749 | 0.900 | 279 794 |
| diagnostic usage 2 | 0.784 | 0.682 | 0.730 | 0.754 | 0.822 | 374 820 |
| diagnostic usage 1 | 0.674 | 0.753 | 0.711 | 0.703 | 0.656 | 491 214 |
| causal interaction 4 | 0.923 | 0.249 | 0.392 | 0.508 | 0.964 | 259 620 |
| causal interaction 3 | 0.897 | 0.324 | 0.476 | 0.545 | 0.934 | 335 838 |
| causal interaction 2 | 0.868 | 0.490 | 0.626 | 0.627 | 0.869 | 456 842 |
| causal interaction 1 | 0.848 | 0.627 | 0.721 | 0.691 | 0.803 | 555 127 |
The results of the procedure were evaluated with respect to precision, recall, F1 score, specificity and accuracy in a 5-fold cross-validation process as shown in the equations (1)–(5). This ensures high-quality data and represents a true upgrade of the BRENDA data. Table 4 displays an overview of the data content and the results of the evaluation procedure.
| (1) |
| (2) |
| (3) |
| (4) |
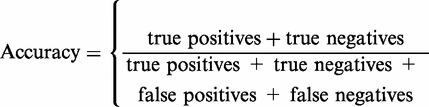 |
(5) |
The integrated biochemical reaction database BKM-react
Information on organism-specific metabolic networks and pathways are essential for many applications including drug target identification, metabolic engineering, etc. Within systems biology genome-wide metabolic models are developed that allow a prediction of cellular reactions to change in the environment or genetic modifications. An essential prerequisite for these applications is a complete set of biochemical reactions for the constructions of the metabolic pathways. BRENDA contains a set of ∼62 000 unique fully characterized enzyme-catalysed reactions (plus ca. 27 000 reactions where, e.g., one of the products was not explicitly determined in the paper). About 10 000 of these are specified in the literature as reactions occurring in the cell. About 8400 reactions are stored in the KEGG Reaction database and MetaCyc stores ∼10 000 reactions.
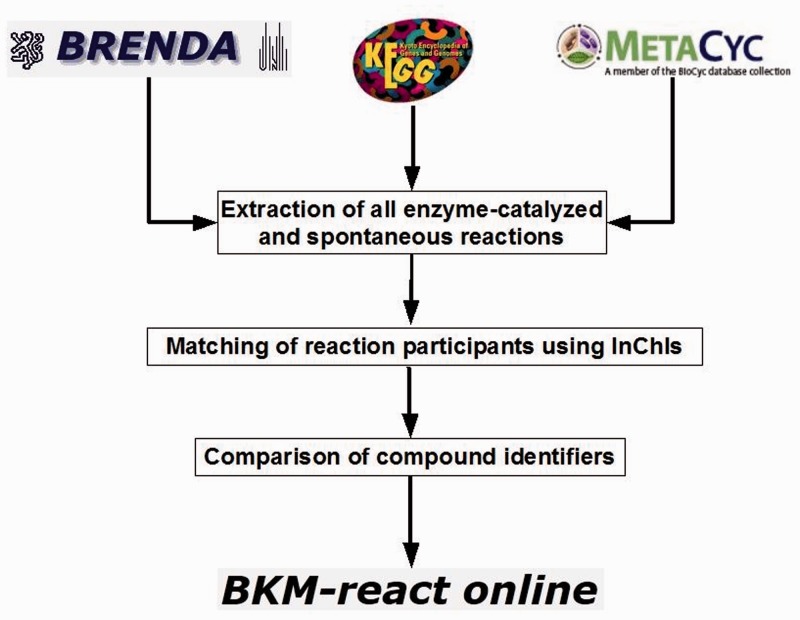
The contents of these databases are overlapping to a considerable degree. Due to the fact that—as in enzymes—a large number of different names are in use for the substrates and products the identity of two reactions is far from obvious. Matching the compounds via their structures has been more promising. The procedure outlined in Figure 3 is based on a first comparison of the InChIs (30) generated from molfiles plus a match with respect to the names and is described in detail in (17). The procedure led to a set of 2890 reactions (15%) common to all databases. 10% occur in two databases and the rest are unique to one of the databases.
Figure 3.
Workflow for matching reactions and compounds in BKM-react.
The often implicitly defined stereochemistry proves to be the major problem. If one of the stereoisomers is much more frequently found than the other the stereodescriptors are often omitted (e.g. the L in L-methionine or the D in D-glucose). The databases follow different strategies to cope with that problem. Assigning the α- or β-anomeric form in carbohydrates is another difficulty.
BKM-react can be accessed via the BRENDA website or with an independent URL. The query system allows searches via EC numbers, pathways, substrate or product names, or identifiers. Besides showing the reactants the result page also lists the stoichiometry and unbalanced reactions.
The enzyme detector
Several databases provide functional annotations of genomes either based on different computational methods or on hand-curated annotations. An initial comparison of the main annotation hosts for nine different prokaryotic organisms revealed 70% inconsistencies. Therefore, we implemented the annotation pipeline EnzymeDetector. This tool automatically compares and evaluates the assigned enzyme functions from the main annotation databases NCBI, KEGG, PEDANT (31,32), Pseudomonas Genome Database V2 (33) and SwissProt. The obtained data are supplemented with our own developed function prediction. This is based on a sequence similarity analysis, on manually created organism-specific enzyme information from BRENDA, and on sequence pattern searches (34). With these integrated data the user can estimate the reliability of the found annotation predictions. A customizable scoring scheme was developed by comparing the annotations found in the different sources to the manually curated data and to the annotations found in SwissProt.
All data found in the several integrated sources are stored in a database and can be accessed on the web interface of the program. Results can be viewed in different ways:
The tabular view shows the results sorted by gene identifier. They can be sorted by EC number or accepted enzyme name. The search results can be downloaded as a csv file.
The statistics view shows a statistical evaluation of the results. It is possible to chose between a static view applying the default settings or a dynamic view which applies the user-chosen constraints.
The annotation comparison view allows the comparison of the enzymes of the selected organisms to the enzymes of one or two other organisms.
The pathway view shows the total number of enzymes, the found and the missing enzymes in the pathways of KEGG and MetaCyc.
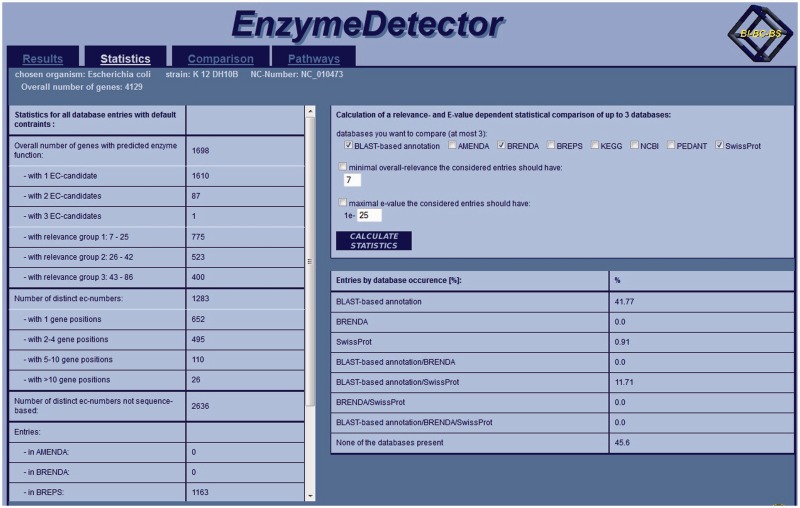
Figure 4 shows a statistical view of the search results for Escherichia coli strain: K-12 DH10B. The database is updated bi-annually.
Figure 4.
Statistical view of the search results for E. coli K-12 DH10B in EnzymeDetector.
THE BRENDA PORTAL
Searches for enzymes and enzyme data can be performed in multiple ways starting from the website.
The Quick Search provides direct searches for all of the ∼50 information fields as well as a full-text search. The latter includes a search in all commentary sections of the database which are not separately listed on the website.
The Advanced Search offers the possibility to combine many search criteria and search in a target-oriented manner thus avoiding overlong lists of results.
The Protein-specific Search opens a window for searching with UniProt sequence IDs or with protein sequences.
The Genome Explorer displays enzymes on genomes and can also be used to compare enzyme localizations in different organisms.
The Substructure Search provides a tool for drawing molecules or fragments thereof. This gives access to the BRENDA ‘Ligands’ which comprise a large quantity of molecules that interact with enzymes such as substrates, products, inhibitors, etc. Ligands can also be accessed using their names and searching with Quick Search → Ligand.
The Taxonomic Tree shows all organisms stored in the respective NCBI database and provides links to the BRENDA enzyme entries. Organisms as enzyme sources can also be searched using Quick Search → Organism.
The EC Explorer gives access to the hierarchical assembly of EC classes in a clearly arranged display. Enzyme data can be accessed from here as well.
The Ontology Explorer is the entry portal for a variety of anatomic, chemical or other ontologies. Enzymes connected with any of the terms can be displayed.
ACCESSIBILITY
The BRENDA web sites are freely accessible at http://www.brenda-enzymes.org. The manually annotated BRENDA data can be downloaded as a single text file at the website (http://www.brenda-enzymes.org/brenda_download/index.php).
License Information can be viewed at http://www .brenda-enzymes.org/index.php4?page=information/copy.php4.
Computer-based access to BRENDA is possible via SOAP (http://www.brenda-enzymes.org/soap2)
Enzyme kinetic data can be obtained via an SBML output.
FUNDING
European Union: Serving Life-science Information for the Next Generation (SLING) [226073]. Funding for open access charge: SLING [226073].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to express their thanks to all collaborating scientists, who performed the literature annotation and created the ligand structures.
REFERENCES
- 1.Gremse M, Chang A, Schomburg I, Grote A, Scheer M, Ebeling C, Schomburg D. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme source. Nucleic Acids Res. 2011;39:D507–D513. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009 – an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Matos P, Adams N, Hastings J, Moreno P, Steinbeck C. A database for chemical proteomics: ChEBI. Methods Mol. Biol. 2012;803:273–296. doi: 10.1007/978-1-61779-364-6_19. [DOI] [PubMed] [Google Scholar]
- 4.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald AG, Boyce S, Tipton KF. ExplorEnz: the primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009;37:D593–D597. doi: 10.1093/nar/gkn582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liébecq C. IUPAC-IUBMB Joint Commission on Biochemical Nomenclature (JCBN) and Nomenclature Committee of IUBMB (NC-IUBMB) Biochem. Mol. Biol. Int. 1997;43:1151–1156. [PubMed] [Google Scholar]
- 7.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Söhngen C, Chang A, Schomburg D. Development of a classification scheme for disease-related enzyme information. BMC Bioinform. 2011;12:329. doi: 10.1186/1471-2105-12-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotera M, Hirakawa M, Tokimatsu T, Goto S, Kanehisa M. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol. Biol. 2012;802:19–39. doi: 10.1007/978-1-61779-400-1_2. [DOI] [PubMed] [Google Scholar]
- 16.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang M, Stelzer M, Schomburg D. BKM-react, an integrated biochemical reaction database. BMC Biochem. 2011;12:42. doi: 10.1186/1471-2091-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quester S, Schomburg D. EnzymeDetector: an integrated enzyme function prediction tool and database. BMC Bioinform. 2011;12:376. doi: 10.1186/1471-2105-12-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, Bairoch A, Schomburg D, Tipton KF, Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004;32:D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin DM, Miranda-Saavedra D, Barton GJ. Kinomer v. 1.0: a database of systematically classified eukaryotic protein kinases. Nucleic Acids Res. 2009;37:D244–D250. doi: 10.1093/nar/gkn834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herráez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 27.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnhammer ElL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 29.Heinen S, Thielen B, Schomburg D. KID – an algorithm for fast and efficient text mining used to automatically generate a database containing kinetic information of enzymes. BMC Bioinform. 2010;11:375. doi: 10.1186/1471-2105-11-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein SE, Heller SR, Tchekhovskoi D. Proceedings of the 2003 International Chemical Information Conference. Infonortics: Nimes; 2003. An open standard for chemical structure representation: the IUPAC chemical identifier; pp. 131–143. [Google Scholar]
- 31.Frishman D, Mokrejs M, Kosykh D, Kastenmüller G, Kolesov G, Zubrzycki I, Gruber C, Geier B, Kaps A, Albermann K, et al. The PEDANT genome database. Nucleic Acids Res. 2003;31:D207–D211. doi: 10.1093/nar/gkg005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter MC, Rattei T, Arnold R, Güldener U, Münsterkötter M, Nenova K, Kastenmüller G, Tischler P, Wölling A, Volz A, et al. PEDANT covers all complete RefSeq genomes. Nucleic Acids Res. 2009;37:D408–D411. doi: 10.1093/nar/gkn749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock REW, Brinkman FSL. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009;37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannert C, Welfle A, Aus dem Spring C, Schomburg D. BrEPS: a flexible and automatic protocol to compute enzyme-specific sequence profiles for functional annotation. BMC Bioinform. 2010;11:589. doi: 10.1186/1471-2105-11-589. [DOI] [PMC free article] [PubMed] [Google Scholar]