Abstract
A novel resource centre for TP53 mutations and mutants has been developed (http://p53.fr). TP53 gene dysfunction can be found in the majority of human cancer types. The potential use of TP53 mutation as a biomarker for clinical studies or exposome analysis has led to the publication of thousands of reports describing the TP53 gene status in >10 000 tumours. The UMD TP53 mutation database was created in 1990 and has been regularly updated. The 2012 release of the database has been carefully curated, and all suspicious reports have been eliminated. It is available either as a flat file that can be easily manipulated or as novel multi-platform analytical software that has been designed to analyse various aspects of TP53 mutations. Several tools to ascertain TP53 mutations are also available for download. We have developed TP53MULTLoad, a manually curated database providing comprehensive details on the properties of 2549 missense TP53 mutants. More than 100 000 entries have been arranged in 39 different activity fields, such as change of transactivation on various promoters, apoptosis or growth arrest. For several hot spot mutants, multiple gain of function activities are also included. The database can be easily browsed via a graphical user interface.
INTRODUCTION
TP53 gene mutation is the most frequent genetic alteration found in human cancer. Since the first discovery of TP53 alterations in 1989, >35 000 mutations have been described in various types of tumours (Supplementary Figure S1). One of the greatest contributions to the study of TP53 mutations has been provided by molecular epidemiology and its applications. We will not discuss these epidemiological studies in detail, as they have been the subject of many detailed reviews and are summarized in Supplementary Table S1 (1,2). These studies demonstrate a link between exposure to various types of carcinogens, specific mutational events in the TP53 gene and the development of specific cancers. The most striking example is that of tandem mutations, specifically induced by ultraviolet radiation, which are only observed in skin cancers. The relationship between G->T transversion and lung cancer in smokers or the mutation of codon 249 observed in aflatoxin B1-induced liver cancers is also demonstrative. More recently, a specific pattern of mutational events in the TP53 gene has been observed in nephropathy-associated tumours that support the involvement of aristolochic acid, a potent mutagen found in traditional medicines, in the aetiology of this cancer. These studies were made possible by the fact that TP53 is the only gene that combines several specific features used to study the origin of carcinogenesis in a human population: (i) it is mutated in many types of cancers; (ii) the mutation frequency is high; (iii) the gene is predominantly modified by point mutations; and (iv) the gene is small enough to be relatively easy to analyse. Whole genome sequencing analysis of multiple types of cancer has confirmed these studies, as the mutation spectra were found to be similar for the TP53 gene and for the entire genome (3).
TP53 mutation databases have also been a framework for many studies on the structure–function relationship of the TP53 protein and the design of novel drugs that can target specific mutant TP53 (4). The first issue of the Universal Mutation Database (UMD) TP53 mutation database was published in 1992 (5) with 360 mutations and has been regularly updated over the years (Supplementary Figure S1) (6–11). Since the last report published in the NAR Database Issue in 1998 (12), the UMD TP53 mutation database has been extended to include functional data of mutant TP53 (9–11). Several tools to analyse and assess TP53 mutation accuracy have also been developed. In 2012, a new website has been designed to host the latest release of the UMD TP53 database (http://p53.fr). Using specific statistical analysis, this database has been highly curated to remove artefactual data (13). It is available for download with several novel analytical tools that will allow the user to perform multiple types of analysis. The mutation database has been linked to a novel database of mutant TP53 activity comprising >100 000 entries related to the multiple characteristics of mutant TP53. Making all of these tools available at a single website provides a useful way to perform multiple analyses on mutant TP53. This integration of functional and structural data with cancer mutations will lead to a novel type of Locus-Specific DataBase (LSDB) that will be more versatile and more valuable than single mutation repositories.
EVOLUTION OF THE TP53 MUTATION DATABASE
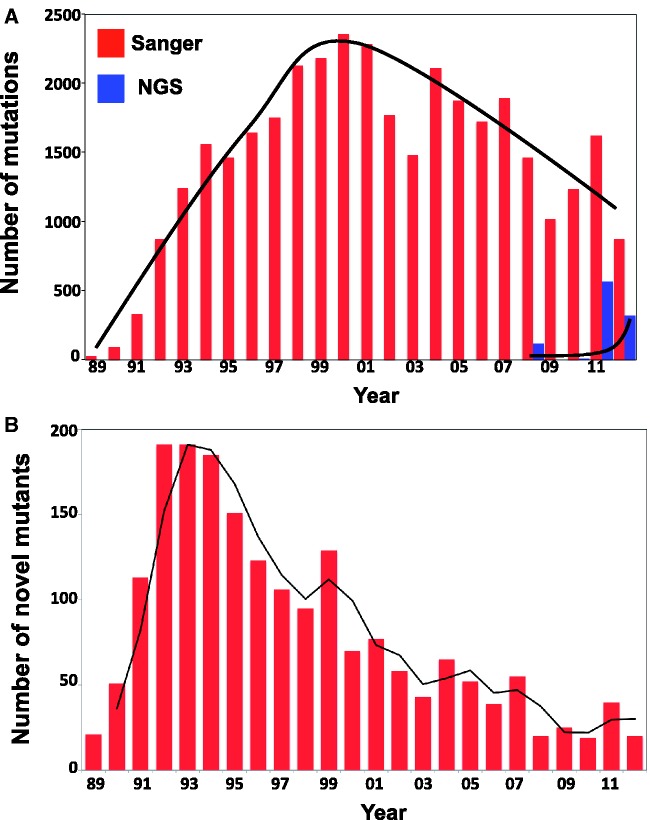
The trend of publications describing TP53 mutation follows a biphasic curve (Figure 1A). After a decade of a regularly increasing number of publications, the number of reports has now slowed down (Figure 1A). This decrease is not owing to lack of interest in TP53, but to the difficulty of publishing TP53 mutations in peer-reviewed journals owing to their lack of novelty. In recent publications, TP53 mutations are not fully described owing to journal space considerations. They are either listed as supplementary materials or, increasingly frequently, not described at all. This problem is not specific to TP53 and applies to many other genes, raising an important issue related to the publication of somatic mutations and the future growth of LSDB. Since 2008, with the development of Next Generation Sequencing (NGS) and the various publications of tumour genome sequences, a new growth of reports describing TP53 mutations has been observed.
Figure 1.
Trends in TP53 mutation publications. (A) Number of mutations published each year using either Sanger methodology (Red) or NGS (blue). For 2012 (last column on the left), only the first 6 months of the year were taken into account. Only publications describing molecular analysis of TP53 mutations were included in this analysis. (B) Number of novel TP53 mutants described each year since the first discovery.
The rate of description of novel TP53 mutants is also biphasic (Figure 1B) (In this review, we will make a clear distinction between TP53 mutations corresponding to entries in the database and TP53 mutants corresponding to the protein variants encoded by the mutations: e.g. the database contains 1530 mutations that encode for the R175H mutant). It increased exponentially for several years and, in 1995, >50% of TP53 mutations had already been discovered. The description rate subsequently slowed, to reach a plateau corresponding to a saturation limit, at which most TP53 mutants associated with loss of tumour protection activity have been identified.
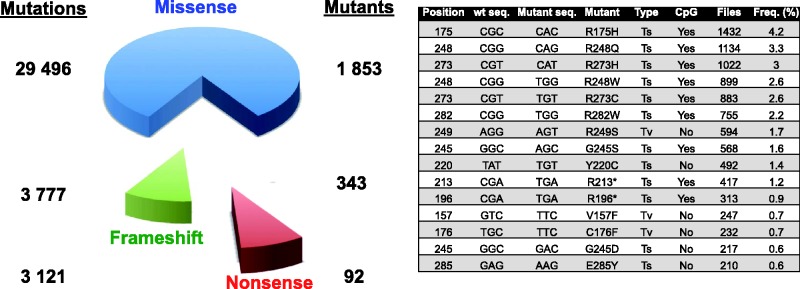
Compared with other tumour suppressor genes, the mode of inactivation of TP53 is unique. Most tumour suppressor genes are inactivated by frameshift or nonsense mutations leading to absence of protein synthesis (or production of a truncated product), whereas >80% of TP53 alterations are missense mutations that lead to the synthesis of a full-length protein that accumulates in the nucleus of the tumour cell (Figure 2). The latest issue of the database includes 1853 different TP53 variants derived from missense mutations (Figure 2). This selection to maintain mutant TP53 in tumour cells is necessary for both a dominant negative activity towards wild-type (wt) TP53 expressed by the remaining allele, and for a gain of function that transforms mutant TP53 into a dominant oncogene (see below). On the other hand, compared with other oncogenes that display mutations that are restricted in a few hot spots associated with a gain of function, TP53 missense mutations are scattered throughout the protein (human TP53: 393 aa). Each residue of the TP53 protein from codon 50 to 360 has been found to be mutated at least once in human tumours (human TP53: 393 aa). Although a few hot spots can be observed, they do not exceed 8–10% and vary considerably in different types of cancer (Figure 2 and Supplementary Table S2). In several common cancers, such as lung and breast cancer, no mutant represents >5% of all mutations (Supplementary Table S2).
Figure 2.
Summary of the content of the latest version of the TP53 mutation database. Mutants and mutations have been classified into three categories (missense, frameshift and nonsense). The features of the 15 most frequent mutations in the database are shown in the right part of the figure.
It has been assumed that the majority of TP53 mutations are localized in exons 5–8 that encode the TP53 DNA-binding domain. This assumption has led to a large bias in the strategy used for TP53 mutation analysis in tumours, as >80% of these studies have focused on this region. The UMD TP53 database comprises the strategy used by the authors for sequencing analysis. The latest release of the TP53 mutation database includes several novel studies that cover the entire TP53 gene using either conventional Sanger sequencing or NGS. Results obtained with both approaches display a similar distribution of TP53 mutations, with 15% localized in exons 4, 9 and 10, indicating that the entire TP53 gene must be screened to correctly assess the TP53 status of the tumour (Supplementary Figure S2A). Analysis of the latest release of the database also reveals that frameshift mutations are statistically more frequent in exons 4 and 9 than in exons 5–8 (P < 0.001), but the origin of this difference is unknown (Supplementary Figure S2B).
It is interesting to note that all new TP53 mutants reported in 2012 are localized outside exons 5–8 in studies using whole genome sequencing via NGS.
QUALITY OF THE DATABASE AND CURATION
One of the most important features for any type of database is the quality of the data. Mutation databases are known to be polluted by spurious mutations predominantly derived from DNA sequencing artefacts (14,15). As early as 2001, the quality of various studies was questioned (16). Inaccuracy of the TP53 mutation database can have multiple consequences. First, it can induce a bias in the interpretation of epidemiological or clinical analysis. Second, the TP53 mutation database is used by multiple programmes as a trainer set for the development of algorithms used for the prediction of deleteriousness of protein mutation. Artefactual mutations would be detrimental to the development of these tools.
In 2003, C. Ishioka and coworkers described the construction of >2000 TP53 mutants, and the biological activity of each mutant was evaluated in vitro in a yeast assay (17). This novel data set led to the possibility of performing cross-analysis between the activity of the various mutants and their occurrence in the database. The two data sets have been merged, and we have shown that mutants frequently described in the literature are true mutants that display loss of function (18,19). However, >50% of the rare TP53 mutants displayed an activity that could not be distinguished from that of wt TP53. These particular mutations have been frequently found in studies reporting multiple mutations in one tumour, silent mutations or lacking mutation hotspots. The majority of these reports were associated with particular methodologies, such as nested PCR on DNA extracted from paraffin-embedded tissue, for which key controls were not satisfactory (18). These analyses raise a number of concerns about analysis of TP53 mutations and indicate the need for caution in global analysis of TP53 mutations from the database. These studies led to the proposal of several recommendations for rigorous analysis of TP53 status in human tumours and the first release of a curated TP53 database with the removal of artefactual publications (20).
Recently, a more accurate curation of the TP53 database has been performed using novel independent criteria [compared with the remaining activity (ACT) that was used previously] to evaluate the quality of the publications in the database (13). These novel criteria are more objective, unrelated to any TP53 function and largely independent of each other. Using data-driven methods, we were able to show that 10% of the publications analysed contain large numbers of artefactual mutations. The current release of the database (2012 R1) is now available in two versions, the curated version that has been purged of outlier studies and a full uncurated version that contains all publications. This latter version can be used to develop more accurate ranking algorithms.
The current release of the UMD database is derived from three different data sets based on the origin of the material used for the study, tumour for somatic mutations, normal tissue for germline mutations and cell lines. For cell lines, the database has also been highly curated, as previous studies have shown that contaminated or incorrectly identified cell lines have led to numerous controversial observations. In particular, the NCI-60 cell line panel contained numerous inconsistencies and controversial results between publications (21). Using the data from the Cancer genome project at the Welcome Trust Sanger Institute hosted at the Catalogue of Somatic Mutations in Cancer (COSMIC) website (22) and manual curation of the literature (21), an accurate database of TP53 alterations in cell lines is now available at the TP53 website.
THE 2012 RELEASE OF THE UMD DATABASE
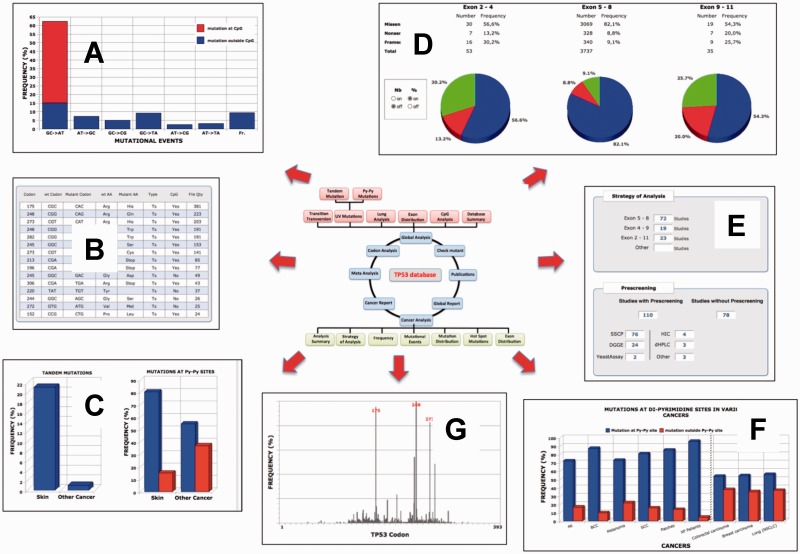
TP53 mutation databases, like other mutation databases, are usually available as a flat file that can be downloaded from websites and processed with any spreadsheet applications. The 2012 issue of the TP53 mutation database has been developed by using novel relational database software and is now provided as a stand-alone application with multiple tools to analyse various aspect of TP53 mutations. This application, TP53_DB_INVEST, includes two main options that allow either global analysis of the entire database or detailed analysis of a specific subset of the database chosen by the user. Up to 20 different types of database analysis can be performed. A graphic user interface allows rapid navigation between the various menus of the software. Results are shown in tabular and/or graphic displays (Figure 3A–G). They can be saved in pdf format or as csv files for further analysis. The various types of analysis are fully described in a 40-page manual with multiple examples. The manual is available both as supplementary material with this article or can be downloaded from the TP53 website (http://p53.fr). Another feature of this software is the possibility for the user to compare his/her own set of TP53 mutants to the TP53 database and export quality tables ready for publication free of any typing errors and codon inaccuracies and using the international nomenclature for mutations. The user only needs to enter three types of information: (i) case ID, (ii) position of the mutation (at the codon level) and (iii) mutant sequence. Data can be entered manually or imported via a txt file. Up to 100 TP53 mutants can be analysed. The software then automatically displays a table comprising all information concerning wt and mutant codons and amino acids (one- and three-letter codes). This information is linked to specific information about the likelihood of each mutation, its frequency in the database and it activity. The nomenclature of the TP53 mutants is based on the guideline described by Den Dunnen and Antonarakis (23). TP53_DBM_INVEST is available for both Mac and Windows environments and can be downloaded from the TP53 website (http://p53.fr).
Figure 3.
An overview of the various features of TP53_DBM_INVEST. (A) Distribution of mutational events; (B) List and description of hot spot mutations; (C) Distribution of tandem mutations; (D) Frequency of TP53 mutations in various regions of the TP53 gene; (E) Summary of the strategy used for the analysis of TP53 mutations (top panel: number of studies focusing on sequencing of different TP53 gene regions; bottom panel: number of studies adopting specific experimental approaches); (F) Frequency of TP53 mutations at dipyrimidine sites for specific cancer types; (G) Overall distribution of TP53 mutations in the protein.
TP53MUTLOAD
The TP53 protein is a tetrameric transcription factor that binds a loose DNA response element (TP53 RE) found in several hundred genes that are differentially activated depending on the cell type, identity, extent of damage and various other parameters that have yet to be identified (24). Although the main function of TP53 is to integrate the diversity of cellular stress responses, more recent information suggests a broader function as a communicator between various physiological processes such as stem cell production and viability, fertility, longevity or metabolism (25,26). This is achieved via various transcription-dependent and -independent activities of the TP53 protein that can act in multiple cellular pathways such as senescence, cell death (via apoptosis or autophagy) or cell cycle progression.
It has now been clearly established that the 2288 TP53 mutants that have been described in human cancer are highly heterogeneous. This complex heterogeneity has multiple clinical consequences (27,28). First, patients with different TP53 mutations in their tumours have different outcomes and can also display different responses to chemotherapy (29). Second, therapeutic drugs targeting mutant TP53 have been shown to be selective for only a subset of mutants. For example, NSC319726 compound has been shown to essentially target mutant R175H whereas PhiKan083 molecule has been developed to specifically restore mutant Y220C (30,31).
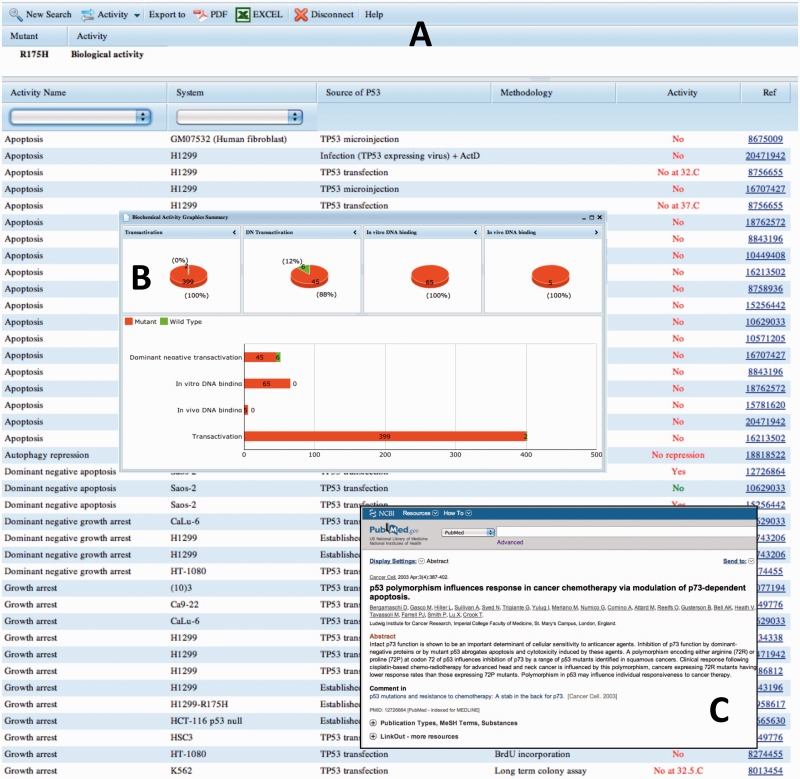
This heterogeneous behaviour of mutant TP53 has been investigated for >20 years, leading to a huge, but disparate, literature. This literature has been included in the new version of the database, TP53 MULTLOAD for TP53 Mutant Loss of Activity Database. TP53 protein properties have been classified into four classes, each including between 4 to 9 specific characteristics, with a total of 26 characteristics (Supplementary Table S3). Database information has been manually curated from the literature. Although all data are experimental results that must be interpreted cautiously when taken individually, the power of the database is due to repetition of multiple analyses performed in various experimental systems. The database contains not only data for mutants found in human cancer, but also all artificial mutants that have been constructed to analyse specific properties of the protein. It notably includes mutants defective for post-translational modifications, oligomerization or nuclear localization. Each file includes specific information on the methodology used for the analysis, such as the recipient cell lines or the source of TP53 used for the analysis. The first issue of this database contains 120 000 entries for 3000 TP53 mutants. The properties of each mutant can be explored via a user-friendly web interface (Figure 4). Results are displayed either as table that can be filtered for a specific subset of data or as a graphic summary. Each file also includes the PMID number of the reference with a hyperlink to the PubMed entry (Figure 4). Results can be exported as a csv file for further analysis by the user. A 50-page manual with several examples is available for download and explains each type of analysis.
Figure 4.
Biological activity report for the hotspot mutant R175H. (A) Downloadable table view listing results from different assays, systems and experimental approaches. (B) Graphical summary of DNA binding and transactivation results. (C) Example of the hyperlink to the publications curated in this section of the database.
TP53_MutAssessor
Predicting the pathogenicity of missense variants is extremely important, as the high throughput of NGS reveals billions of nucleotide variations that must be classified as deleterious or tolerated. Multiple computational methods have been developed to assess the impact of these variations, but most of them use global parameters and do not take into account the specificity of each gene (32).
TP53_MutAssessor is a novel and original tool that defines an ID card for mutant TP53. It displays two types of information. The first type of information is related to functional data, localization of the mutation in the various TP53 isoforms or in the other members of the TP53 family (TP63 and TP73), phylogenetic conservation or structural data that can be used for any type of TP53 mutant i.e. natural mutant found in human cancer or artificial mutant constructed for specific research purposes.
The second type of information is related to statistical analysis of the TP53 mutation database and is more specific to mutants found in human cancer. The software defines a confidence index for each TP53 mutant based on various parameters, such as frequency in the database, loss of activity or association in outlier studies. Cancer type is also an important criterion to be taken into account, as some nucleotide substitutions such as tandem mutations at dipyrimidine sites are much more frequent in skin cancer than in internal tumours. Additionally, exonic mutations that can alter splicing have been documented; this is an important feature, as they are often missed. The strength of these various criteria has been evaluated in a recent analysis of the TP53 database, and they allow clear identification of suspicious mutations (13). TP53 mutant pathogenicity analysis is based on the TP53 functional database used in Mutload. The TP53 Mut Assessor database includes every potential amino acid substitution for each position of the protein (393 residues) whether or not they have been previously described. It also includes nonsense and frameshift mutations. Although only 3000 of the 8000 mutants included in the database have been described in a cancer, this database also includes the properties of artificial mutants in specific domains (transactivation, oligomerization) or signal sequences (nuclear localization, post-translational). Up to 10 TP53 mutants can be analysed simultaneously.
TP53 Mut Assessor is available for both Mac and Windows environments and can be downloaded from the TP53 website. A full documentation is available with the software.
CONCLUSIONS/PERSPECTIVES
The evolution of the UMD TP53 database from a flat file describing 300 mutations in 1992 to an integrative resource centre available from a single website parallels the evolution of our knowledge on TP53 and its importance in cancer research. Studies of the loss and/or gain of activity of TP53 mutants is an active field of investigation in both basic and clinical research and is far from being fully understood. Two series of recent reports have revolutionized the current knowledge on TP53 mutations and their relations to cancer. First, Jackson et al . (33) showed that mice expressing missense TP53 mutants in breast tumours displayed a better response to therapy compared with mice harbouring tumours with a wt allele. Tumours expressing mutant TP53 with and without loss of the wt allele behave differently, as those that retain the wt allele were the less responsive to therapy. Although this observation contradicts the prevailing opinion that TP53 mutations are associated with lack of response to various drugs used in chemotherapy, it supports isolated observations in human breast cancer that describe a better response to therapy for tumours expressing non-functional TP53. Similarly, medullary breast cancers that display a frequency of TP53 mutations of ∼95% are associated with a good prognosis (34). Further investigations will determine whether or not these observations can be extended to other cancer types. A second series of reports showed that TP53 mutants deficient for growth arrest, apoptosis and senescence are still active for tumour suppression (35,36). These studies suggested that other pathways such as metabolic regulation or antioxidant function must be explored to gain a better knowledge on mutant TP53 and its relation to neoplasia. All these studies were performed in mice, and further studies in humans will be required to clarify the true anti-neoplastic function(s) of the TP53 gene. An important issue from the work of Jackson et al. concerns knowledge of the status of the TP53 wt allele in human tumours, which is a complex issue owing to the heterogeneity of the tumour materials. NGS will probably be able to resolve this problem by providing more accurate data. Continuous integration of these data will be essential to provide an accurate picture of this complex protein.
The cancer gene LSDB era will come to an end as data are now collected and compiled from large data coordination centres such as The Cancer Genome Atlas (TCGA) and COSMIC (37). Mutations on thousands of genes can be browsed, but they are not linked to any functional data, which constitute an essential issue as it has become clear that, regardless of the genes considered, each different mutant can be associated with a different ‘penetrance’. The integration of functional and structural data with cancer mutations, as exemplified in this TP53 mutation resource centre, will lead to a novel type of LSDB that will be more valuable and beneficial to the scientific community than single mutation repositories.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–2.
FUNDING
Swedish Cancer Society and the Swedish Research Council (VR) (to T.S.); Associazione Italiana per la Ricerca sul Cancro (AIRC) [IG#5506 to G.F. in part]. Funding for open access charge: Swedish Cancer Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 2.Soussi T. Advances in carcinogenesis: a historical perspective from observational studies to tumor genome sequencing and TP53 mutation spectrum analysis. Biochim. Biophys. Acta. 2011;1816:199–208. doi: 10.1016/j.bbcan.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Soussi T. TP53 Mutations in human cancer: database reassessment and prospects for the next decade. Adv. Cancer Res. 2011;110:107–139. doi: 10.1016/B978-0-12-386469-7.00005-0. [DOI] [PubMed] [Google Scholar]
- 4.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 5.Caron de Fromentel C, Soussi T. TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes Chromosomes Cancer. 1992;4:1–15. doi: 10.1002/gcc.2870040102. [DOI] [PubMed] [Google Scholar]
- 6.Cariello NF, Beroud C, Soussi T. Database and software for the analysis of mutations at the human p53 gene. Nucleic Acids Res. 1994;22:3549–3550. [PMC free article] [PubMed] [Google Scholar]
- 7.Beroud C, Verdier F, Soussi T. p53 gene mutation: software and database. Nucleic Acids Res. 1996;24:147–150. doi: 10.1093/nar/24.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beroud C, Soussi T. p53 and APC gene mutations: software and databases. Nucleic Acids Res. 1997;25:138. doi: 10.1093/nar/25.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soussi T, Dehouche K, Beroud C. p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum. Mutat. 2000;15:105–113. doi: 10.1002/(SICI)1098-1004(200001)15:1<105::AID-HUMU19>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 10.Beroud C, Soussi T. The UMD-p53 database: new mutations and analysis tools. Hum. Mutat. 2003;21:176–181. doi: 10.1002/humu.10187. [DOI] [PubMed] [Google Scholar]
- 11.Hamroun D, Kato S, Ishioka C, Claustres M, Beroud C, Soussi T. The UMD TP53 database and website: update and revisions. Hum. Mutat. 2006;27:14–20. doi: 10.1002/humu.20269. [DOI] [PubMed] [Google Scholar]
- 12.Beroud C, Soussi T. p53 gene mutation: software and database. Nucleic Acids Res. 1998;26:200–204. doi: 10.1093/nar/26.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edlund K, Larsson O, Ameur A, Bunikis I, Gyllensten U, Leroy B, Sundstrom M, Micke P, Botling J, Soussi T. Data-driven unbiased curation of the TP53 tumor suppressor gene mutation database and validation by ultradeep sequencing of human tumors. Proc. Natl Acad. Sci. USA. 2012;109:9551–9556. doi: 10.1073/pnas.1200019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kern SE, Winter JM. Elegance, silence and nonsense in the mutations literature for solid tumors. Cancer Biol. Ther. 2006;5:349–359. doi: 10.4161/cbt.5.4.2551. [DOI] [PubMed] [Google Scholar]
- 15.Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin. Pathol. 2004;4:1. doi: 10.1186/1472-6890-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 17.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl Acad. Sci. USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soussi T, Asselain B, Hamroun D, Kato S, Ishioka C, Claustres M, Beroud C. Meta-analysis of the p53 mutation database for mutant p53 biological activity reveals a methodologic bias in mutation detection. Clin. Cancer Res. 2006;12:62–69. doi: 10.1158/1078-0432.CCR-05-0413. [DOI] [PubMed] [Google Scholar]
- 19.Soussi T, Kato S, Levy PP, Ishioka C. Reassessment of the TP53 mutation database in human disease by data mining with a library of TP53 missense mutations. Hum. Mutat. 2005;25:6–17. doi: 10.1002/humu.20114. [DOI] [PubMed] [Google Scholar]
- 20.Soussi T, Ishioka C, Claustres M, Beroud C. Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nat. Rev. Cancer. 2006;6:83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- 21.Berglind H, Pawitan Y, Kato S, Ishioka C, Soussi T. Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol. Ther. 2008;7:699–708. doi: 10.4161/cbt.7.5.5712. [DOI] [PubMed] [Google Scholar]
- 22.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, Santarius T, Avis T, Barthorpe S, Brackenbury L, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol. Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ. Introduction: the changing directions of p53 research. Genes Cancer. 2011;2:382–384. doi: 10.1177/1947601911413463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane D, Levine A. P53 research: the past thirty years and the next thirty years. Cold Spring Harb. Perspect. Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monti P, Ciribilli Y, Jordan J, Menichini P, Umbach DM, Resnick MA, Luzzatto L, Inga A, Fronza G. Transcriptional functionality of germ line p53 mutants influences cancer phenotype. Clin. Cancer Res. 2007;13:3789–3795. doi: 10.1158/1078-0432.CCR-06-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb. Perspect. Biol. 2010;2:a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl Acad. Sci. USA. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 33.Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, Tavana O, Yang P, Manshouri T, Li Y, et al. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Cremoux P, Salomon AV, Liva S, Dendale R, Bouchind'homme B, Martin E, Sastre-Garau X, Magdelenat H, Fourquet A, Soussi T. p53 mutation as a genetic trait of typical medullary breast carcinoma. J. Natl Cancer Inst. 1999;91:641–643. doi: 10.1093/jnci/91.7.641. [DOI] [PubMed] [Google Scholar]
- 35.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]