Abstract
Recently, the attention of the research community has been focused on long non-coding RNAs (lncRNAs) and their physiological/pathological implications. As the number of experiments increase in a rapid rate and transcriptional units are better annotated, databases indexing lncRNA properties and function gradually become essential tools to this process. Aim of DIANA-LncBase (www.microrna.gr/LncBase) is to reinforce researchers’ attempts and unravel microRNA (miRNA)–lncRNA putative functional interactions. This study provides, for the first time, a comprehensive annotation of miRNA targets on lncRNAs. DIANA-LncBase hosts transcriptome-wide experimentally verified and computationally predicted miRNA recognition elements (MREs) on human and mouse lncRNAs. The analysis performed includes an integration of most of the available lncRNA resources, relevant high-throughput HITS-CLIP and PAR-CLIP experimental data as well as state-of-the-art in silico target predictions. The experimentally supported entries available in DIANA-LncBase correspond to >5000 interactions, while the computationally predicted interactions exceed 10 million. DIANA-LncBase hosts detailed information for each miRNA–lncRNA pair, such as external links, graphic plots of transcripts’ genomic location, representation of the binding sites, lncRNA tissue expression as well as MREs conservation and prediction scores.
INTRODUCTION
The annotation of the genome-wide transcriptional repertoire has received significant attention during the past few years. Recent discoveries regarding the numerous biological roles of non-coding RNAs (ncRNAs) highlight the biological significance of these previously ‘overlooked’ RNA species. ncRNAs are now considered a biological hotspot; involved in a plethora of cellular processes including either cis- or trans-regulation of protein-coding genes and alternative splicing (1). Many ncRNA families, such as microRNAs (miRNAs) and, more recently, long ncRNAs (lncRNAs) are being vigorously studied for their physiological and pathological implications.
Generally, RNAs longer than 200 nt not possessing a clearly defined open reading frame are characterized as lncRNAs (2). As relevant research progressed, these ncRNAs were further subdivided into distinct subcategories and ‘lncRNA’ is now considered a blanket term including sense, antisense, intronic, bidirectional and intergenic transcripts (2,3). Most of lncRNA function still remains to be uncovered and specified through experimental procedures. However, recent findings have revealed mechanisms involving lncRNAs in diverse cellular processes, such as chromatin remodeling; structural scaffolding of nuclear protein substructures; cell cycle regulation and they have recently been found to play key roles in the circuitry controlling embryonic stem cell state (4–6). Moreover, they may also perform cis-regulatory actions, as they are usually located near protein-coding genes (7), while, putative direct interactions of lncRNAs with protein-coding genes and other RNA molecules have already been suggested (1,3). For instance, lncRNAs have been shown to strongly associate with p53; target RNA polymerase II in both human and mouse (8); bind to Polycomb repressive complexes (1) and even interact with miRNA molecules and regulate gene expression.
miRNAs are small endogenous RNA molecules (∼22 nt) playing a major role in gene expression. They are considered as post-transcriptional gene regulators enabling translational repression, mRNA degradation and gene silencing (9). They target protein-coding genes usually by partial or complete base pairing on specific miRNA recognition elements (MREs) on mRNA sequences (10). More precisely, miRNA target gene interactions usually require binding of 6–8 nt in the so-called seed region (9) of the miRNA 5′-end. miRNAs have been primarily detected to effectively target specific mRNA 3′-untranslated regions (3′-UTRs), on usually highly conserved MREs (11). Recent findings also exhibited functional bona fide miRNA interactions with MREs located in the 5′-UTR region as well as within the coding sequence (CDS) (12).
In a recent study by Huarte et al. (13), a lincRNA-named lincRNA-p21 was found to repress p53 apoptotic function through binding to hnRNP-K. Bozzoni and coworkers (14) suggested a dual function of lncRNAs: a fraction of the transcripts remains in the nucleus as potential pri-miRNAs, while others move to the cytoplasm as functional lncRNAs. The authors have also identified a muscle-specific lincRNA (linc-MD1) targeted by two miRNAs (miR-133 and miR-135). This lncRNA acts possibly as a ‘sponge’, attracting miRNAs targeting two key transcription factors involved in muscle differentiation, and therefore indirectly regulates their expression. Other researchers also support this lncRNA ‘sponge’/‘decoy’ function, reducing the regulatory effect of miRNAs on targeted mRNAs (14,15), miRNA precursors (16,17) or host genes for miRNAs (18). Identification of the underlying links between lncRNA and miRNA families will provide new insights in molecular biology.
In a recent study, Jeggari et al. (19) have identified putative miRNA-binding sites across all annotated human transcripts of GENCODE v11 release, which also includes 10 419 lncRNAs. The authors provided access to these in silico predicted miRNA sites through the ‘miRcode’ web interface. miRcode supports seed-related information; genomic location, binding type, percentage of evolutionary conservation across primates/non-primate mammals/non-mammalian vertebrates as well as possible overlaps with repeat sequences. The implemented prediction pipeline has been based on a seed complementarity algorithm and on TargetScan 6 miRNA seed family nomenclature. However, miRcode includes limited MRE-related information and only on a small fraction of the publicly available annotated human lncRNAs. miRNA target predictions for other species as well as experimentally verified binding sites on lncRNAs are not supported.
Compared with relevant existing studies, DIANA-LncBase provides an extensive amount of predicted miRNA targets on the largest available set of human lncRNAs. Moreover, it offers for the first time a comprehensive collection of computationally predicted MREs on mouse lncRNAs, as well as miRNA–lncRNA interactions supported by experimental data for both human and mouse species. miRNA targets of large collections of mouse lncRNA transcripts have not yet been extensively studied and there are only few lncRNA–miRNA interactions reported in the available literature. The analysis performed includes all available lncRNA data resources in human and mouse, identification of experimentally verified miRNA targets with the use of high-throughput PAR-CLIP and HITS-CLIP experiments (12,20) as well as in silico target prediction using a state-of-the-art algorithm DIANA-microT-CDS (21).
METHODS AND RESULTS
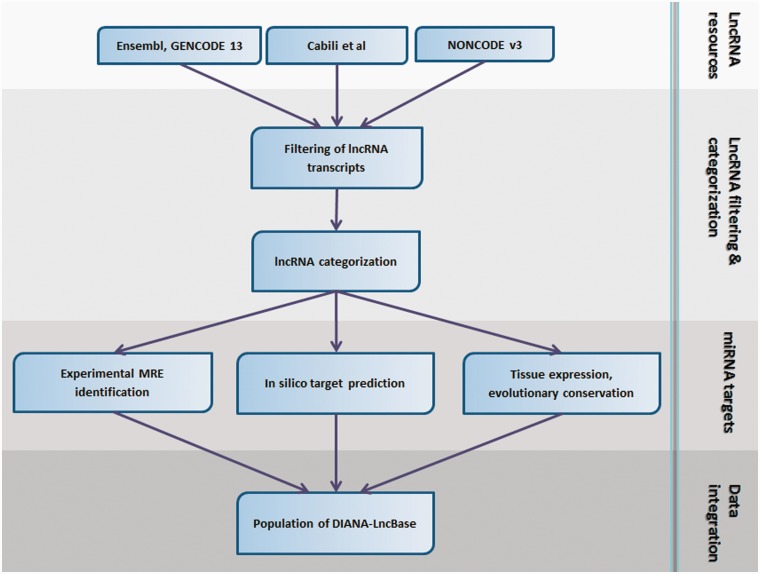
The analysis pipeline, described in the following sections, can be divided into the following distinct steps: collection of lncRNA resources and data pre-processing; miRNA target identification; integration of database entries and population of DIANA-LncBase. An overview of the analysis pipeline is depicted in Figure 1.
Figure 1.
DIANA-LncBase analysis pipeline. The best available lncRNA resources have been collected for human and mouse species. Transcripts shorter than 200 nt as well as transcripts presenting high similarity (>90% overlap) have been removed. lncRNAs have been subsequently categorized in sense, antisense, bidirectional and intergenic with the use of a protein-coding reference set consisting of UCSC and Ensembl genes. Additionally, MREs on lncRNAs have been experimentally verified with the use of high-throughput HITS/PAR-CLIP data and in silico predicted with a state-of-the-art algorithm, DIANA-microT-CDS. Integration of miRNA targets, as well as other lncRNA/miRNA-related information, such as transcripts tissue expression and MREs evolutionary conservation, has been the final step for DIANA-LncBase population.
lncRNA datasets
The analysed lncRNA datasets were formed by combining the best available sources of lncRNA sequences. The integrative dataset of >12 000 lncRNAs from GENCODE 13 (22), and a set of 8195 lncRNAs presented by Cabili et al. (23) were used as the foundation of our collection for human transcripts. The GENCODE dataset provides a high-quality catalog corresponding to the largest set of manually curated lncRNAs. Furthermore, the Cabili et al. dataset is an integration of already annotated and novel lncRNAs from ∼4 billion RNA-Seq reads across 24 tissues. The third source of lncRNA data has been the NONCODE v.3 database (24), supporting a unified set of non-coding transcripts derived from all acknowledged relevant databases and the available literature. The NONCODE lncRNA dataset consists of 33 829 transcripts for Homo sapiens and 37 049 transcripts for Mus musculus. Our mouse dataset is based on the combination of Ensembl 65 and NONCODE v3 mouse lncRNA available sequences.
lncRNAs with high sequence similarity have been removed. In cases of transcript pairs with >90% overlap, the longest lncRNA sequence has been retained. Transcripts shorter than 200 nt were filtered out. Our final human dataset consists of 27 164 lncRNAs, whereas the mouse dataset, slightly larger, is composed of 28 946 lncRNAs.
A reference protein-coding dataset consisting of all Ensembl and UCSC protein-coding genes was utilized, in order to distribute the lncRNAs in four distinct categories:
Sense or antisense lncRNAs overlapping non-intronic parts of protein-coding genes, located in the same or opposite strand (13 578 transcripts).
Bidirectional lncRNAs (2) transcribed in ‘head to head’ orientation and located in close proximity with a protein-coding gene (920 transcripts).
Intergenic lncRNAs located exclusively within intergenic regions (12 666 transcripts).
Additional details concerning the categorization of both human and mouse lncRNA datasets are provided in Supplementary Table S1.
High-throughput data
The exact identification and localization of miRNA-binding sites on target mRNAs can be significantly facilitated by high-throughput RNA sequencing (12,20). Chi et al. identified functional Argonaute (Ago) protein–RNA complexes in P13 mouse brain, using protein crosslinking and immunoprecipitation (HITS-CLIP), while Hafner et al. determined transcriptome-wide-binding sites of RNA-binding proteins and miRNA-containing ribonucleoprotein complexes (miRNPs) utilizing a cell-based crosslinking approach (PAR-CLIP).
PAR-CLIP data
The PAR-CLIP dataset was downloaded from the supplementary material of Hafner et al. (12). It comprises of 17 319 peaks, obtained from all PAR-CLIP AGO experiments throughout the human transcriptome. The T- to C-transition site information for each peak has been considered essential in determining the exact location of miRNA-seed and the mRNA or lncRNA interaction sites.
HITS-CLIP data
The HITS-CLIP dataset was downloaded from the supplementary material of Chi et al. (20). In order to identify miRNA target sites in mouse lncRNAs, raw HITS-CLIP tags from P13 mouse brain have been incorporated in the analysis. HITS-CLIP data have been filtered to retain only those tags overlapping with lncRNA genomic locations. Peak regions have been formed by merging overlapping tags.
miRNA target identification
Experimental MRE identification
Peaks derived from the CLIP experiments suggest direct interaction with miRNAs. Further processing of the CLIP reads is considered essential in order to identify miRNAs binding to the relevant peaks. Therefore, each peak has been further processed by an in-house developed dynamic programming algorithm for MRE identification. The implemented algorithm slides a 9-nt-long window along each transcript and identifies the best possible alignment with the miRNA ‘extended’ seed (nucleotides 1–9 on the miRNA 5′-end). This procedure can detect different binding types, ranging from 4mer to 9mer. Subsequently, MREs related to the top expressed miRNAs in the CLIP experiments have been retained for further analysis. In the PAR-CLIP analysis, putative MREs have been filtered to keep only those located closer than 5 nt to the T to C mutation. Other important parameters used for the final MRE selection for both experiments are the binding type and the binding free energy as calculated by RNAhybrid (25).
In silico MRE identification
The in silico computations were performed using the latest version of the microT algorithm, DIANA-microT-CDS, which is considered as one of the best available miRNA target prediction programs in terms of sensitivity and specificity (21). The target prediction analysis was elaborated for all miRNAs deposited in miRBase v18 (26).
The database
DIANA-LncBase has been designed to accommodate all experimentally verified and in silico predicted miRNA–lncRNA interactions. The database is composed of two distinct modules: one to explore computationally predicted MREs of DIANA-microT-CDS and one to explore experimentally verified target sites. The database interface is specifically designed to facilitate user interaction and database querying.
miRNA–lncRNA interaction data
The database has been populated with entries derived from experimental and in silico MRE identification. A total of 1793 unique miRNA–lncRNA interactions have been identified from the analysis of PAR-CLIP experimental data and the relevant literature, for human, whereas the corresponding number for mouse increases to 3370 interactions using the HITS-CLIP datasets. The in silico dataset hosts >10 million interactions between 56 097 lncRNAs and 3078 miRNAs, for human and mouse.
Tissue expression data
Tissue expression for human lncRNAs has been acquired from two data sources. lncRNAs, derived from Cabili et al., are accompanied with expression values across 24 human tissues. We have also utilized expression patterns for Ensembl lncRNA transcripts, available in the study of Gibb et al. (27). The authors analyzed 72 SAGE libraries in normal tissues and cataloged lncRNAs abundance over a spectrum of 19 normal tissues. For mouse, FANTOM2 database was used to acquire expression for lncRNA transcripts in 20 tissues (28). Targeted lncRNAs in human have been found to be predominantly expressed in testes, brain and lymph node, whereas mouse lncRNAs presented higher abundance in cerebellum, brain and adipose tissues.
Conservation data
In miRNA target prediction, evolutionary conservation is usually a strong indication of target functionality. DIANA-LncBase includes information concerning the conservation of the computationally predicted human and mouse MREs in other species. For the identification of conserved MREs, multiple sequence alignments on 16 vertebrate species were utilized. An MRE is categorized as conserved in another species if it has exactly the same seed sequence. It has been observed that predicted MREs in sense lncRNAs and protein-coding transcripts exhibit similar conservation levels, significantly higher than the rest lncRNA categories, in most of the investigated vertebrate species (Supplementary Figures S2 and S3).
Database search
DIANA-LncBase provides an intuitive and user-friendly interface offering two distinct modules for querying the database. Users can browse the results by querying with a specific miRNA, gene identifier or a combination of the previous terms. Moreover, the target prediction module offers the option to query using a specific genomic location. The provided results of this specialized query correspond to all the predicted lncRNA–miRNA interactions having at least one MRE within the examined location. An additional option is the filtering of computationally predicted target results on lncRNAs. The user can filter the results using an arbitrary microT threshold, fine-tuning prediction sensitivity and precision levels as well as specific lncRNA tissue expression (Figure 3). On the other hand, the experimental module offers access to the experimentally validated targets and hosts an enhanced filtering system, able to reduce results based on species, experimental method, regulation type, validation type, data source, publication year and predicted interaction score (Figure 2). Further details concerning the database search and information regarding the interface of these modules are provided in the help section of the DIANA-LncBase.
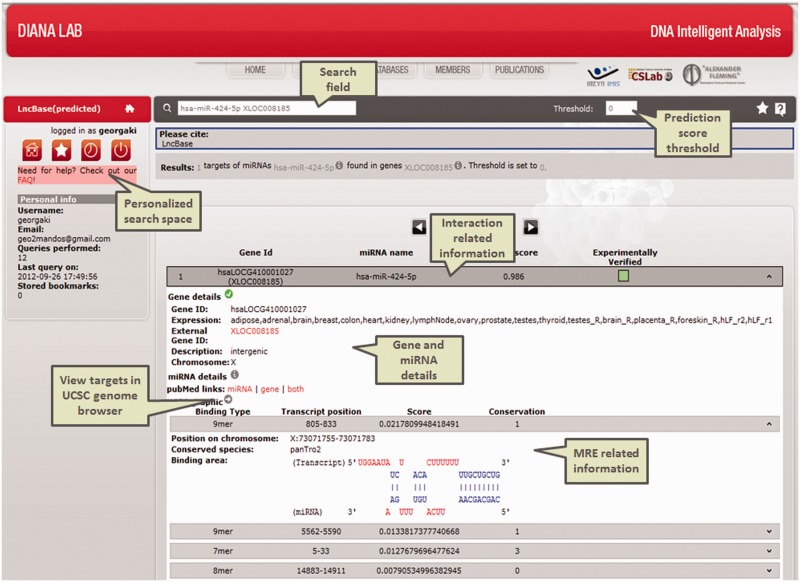
Figure 3.
Screenshot of the predicted module in DIANA-LncBase interface. The results of the query regarding a specific miRNA–lncRNA interacting pair are depicted in the form of an expandable list.
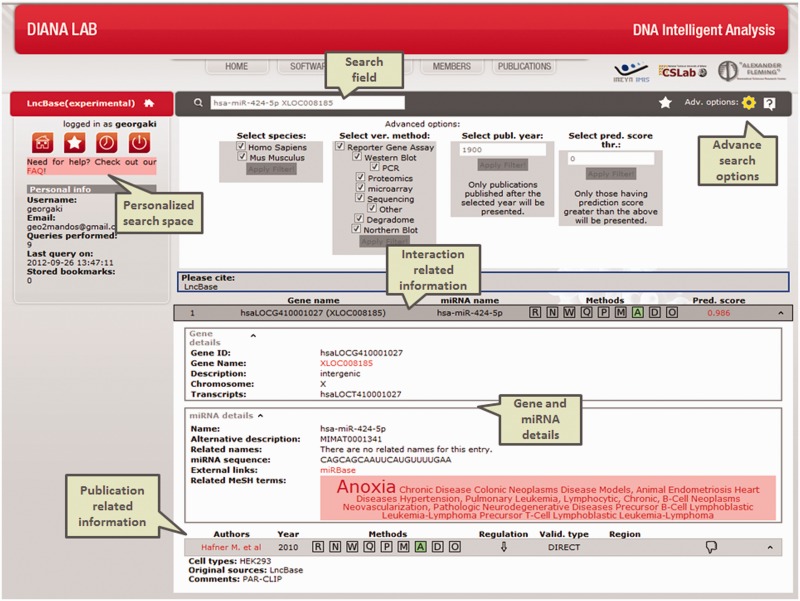
Figure 2.
Results of a submitted query in DIANA-LncBase experimental module. The interface presents information regarding the specified miRNA–lncRNA interaction. miRNA and gene-related information, as well as the advanced search options have been expanded. This interaction supported by the PAR-CLIP high-throughput data is also predicted from DIANA-microT-CDS.
Statistics
High-throughput data analysis and in silico target prediction have also been performed for protein-coding genes. The protein-coding dataset has been formed by combining CDS and 3′-UTR spliced regions of the corresponding mRNA transcripts from Ensembl version 65 (29). Analysis of the experimentally verified targets datasets revealed significantly less miRNA targets sites in lncRNA transcripts, compared with protein-coding genes. More precisely, a total of 1785 and 3274 peaks with MREs have been identified from HITS/PAR-CLIP analyses on human and mouse lncRNAs (Supplementary Tables S2 and S3). The in silico computations, on the other hand, performed for all available miRNAs, have resulted in a high number of MREs in lncRNA transcripts for both species.
The density of the experimentally identified miRNA targets sites per 1000 nt in expressed lncRNA and protein-coding transcripts was calculated using the HITS/PAR-CLIP datasets. Density estimations were also calculated following removal of all sense and protein-coding transcripts with at least one common experimentally verified MRE. In both cases, sense lncRNA transcripts in human, and all lncRNA categories in mouse, present significantly higher MRE densities, compared with protein-coding transcripts (P < 0.05) (Supplementary Figure S1 and Supplementary Tables S4 and S5); an indication that lncRNA targeting by miRNAs might be a more common phenomenon than what we initially expected.
Moreover, we have used multiple sequence alignments in 21 mammal species and assessed the degree of conservation of human lncRNAs and protein-coding exons, using SiPhy (30). The estimated omega conservation scores from the SiPhy algorithm, depicting the local rate of substitutions compared with a neutral tree model, are in agreement with the results of a previous study by Khalil et al. (5). More precisely, the exons on protein-coding transcripts were found more conserved than the exons in lncRNAs across the queried species (Supplementary Figure S4).
CONCLUSION
DIANA-LncBase is a novel database accommodating experimentally verified, as well as in silico predicted lncRNA–miRNA interactions in human and mouse genomes. Since there is a limited number of such interactions reported in the available literature, DIANA-LncBase can provide a significant boost toward understanding the mechanisms of their synergy and regulation. Its maintenance and further expansion are considered essential, in order to keep pace with the exponential growth/annotation of ncRNA transcripts, as well as the rapid distinction of their functions in different cell compartments. DIANA-LncBase aims to develop into a reference repository of lncRNA–miRNA interactions supported from the literature, identified through novel computational approaches, already existing high-throughput data or newly designed Next Generation Sequencing experiments. The outcome of such an approach will provide an unprecedented amount of high-quality data that will become the stepping stone for numerous analyses and future research projects throughout the community.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–4 and Supplementary Methods.
FUNDING
Projects 09 SYN-13-1055 ‘MIKRORNA’ and 09 SYN-13-901 ‘EDGE’ from the Greek General Secretariat for Research and Technology. Funding for open access charge: Project ‘MIKRORNA'.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Ioannis S. Vlachos for his helpful suggestions during the compilation of this work.
REFERENCES
- 1.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA. 2007;13:583–596. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 11.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- 22.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, Xiao H, Liao Q, Luo H, Zhao G, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210–D215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bono H, Yagi K, Kasukawa T, Nikaido I, Tominaga N, Miki R, Mizuno Y, Tomaru Y, Goto H, Nitanda H, et al. Systematic expression profiling of the mouse transcriptome using RIKEN cDNA microarrays. Genome Res. 2003;13:1318–1323. doi: 10.1101/gr.1075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]