Abstract
The International Nucleotide Sequence Database Collaboration (INSDC; http://www.insdc.org), one of the longest-standing global alliances of biological data archives, captures, preserves and provides comprehensive public domain nucleotide sequence information. Three partners of the INSDC work in cooperation to establish formats for data and metadata and protocols that facilitate reliable data submission to their databases and support continual data exchange around the world. In this article, the INSDC current status and update for the year of 2012 are presented. Among discussed items of international collaboration meeting in 2012, BioSample database and changes in submission are described as topics.
INTRODUCTION
For over 30 years, the International Nucleotide Sequence Database Collaboration (INSDC; http://www.insdc.org) has maintained the primary nucleotide sequence database. INSDC has collected nucleotide sequence data and metadata from researchers and has issued the internationally authorized accession number, for data submitters and scientific journals. The INSDC consists of three partners; the DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp/) at the National Institute for Genetics in Mishima, Japan; the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI; http://www.ebi.ac.uk/ena) in Hinxton, UK and the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genbank/) in Bethesda, MD, USA. The INSDC has a uniform policy of free and unrestricted access to the data (1). Under the policy, the INSDC captures, preserves, provides and exchanges the comprehensive nucleotide sequence and associated information on a daily basis. As new sequencing technology has emerged and has been deployed, the scope of sequencing activity has grown enormously, and INSDC has launched new services that deal with the richness of the domain, including repositories for raw data [the Trace Archives for Sanger method and Sequence Read Archive (SRA) for next-generation platforms] (2), assembly data, experimental design details, taxonomic information, functional annotation, project information and sample information. As a traditional data set, assembled sequences and annotations are available from DDBJ (3), the EMBL-Bank component of the European Nucleotide Archive (4) and GenBank from NCBI (5). Routine data exchange, standard formats and the sharing of technology provide global synchrony across the collaboration. In this article, we outline the current status of, and changes to, INSDC including the creation of the BioSample databases (6,7) and some modifications that allow INSDC partners to respond to demands of the research domain.
CONTENT IN 2012
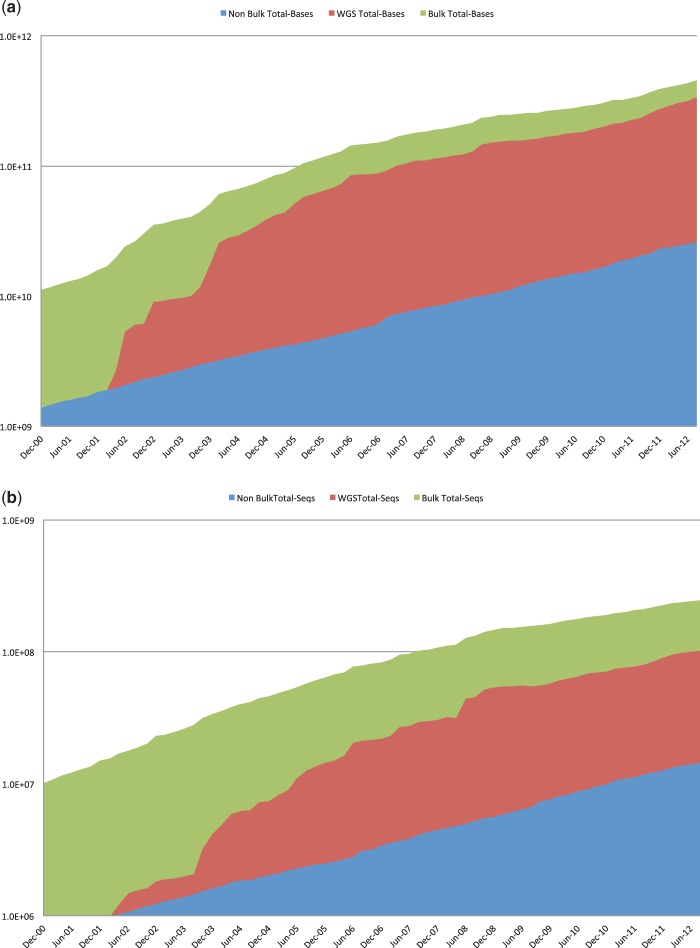
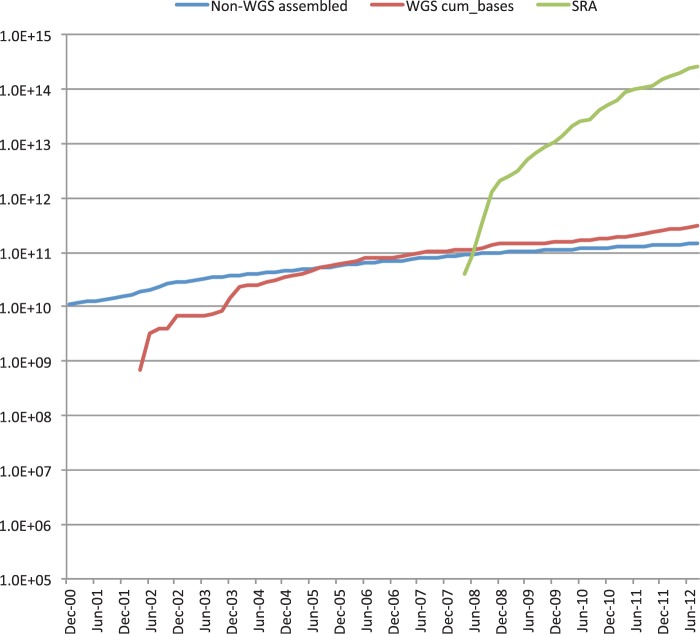
In total, whole INSDC data set has grown overall ∼2-fold in terms of the number of bases in 2012. The latest release 90.0 of DDBJ contains data prepared as of August 24, 2012, from DDBJ, EMBL-Bank/EBI and GenBank, that is, traditional part of INSDC data set. The release consists of 156 952 755 sequence entries and 144 754 534 372 nucleotides. From August 2011 to August 2012, traditional INSDC has grown 1.3-fold in terms of the number of bases and 1.2-fold in the number of entries (Figure 1), whereas SRA for raw data of NGS has grown 2.4-fold in the number of bases (Figure 2).
Figure 1.
Cumulative growth (a) in the number of entry of sequences (b) in the number of nucleotides included in the traditional INSDC sequence archives over time. Bulk sequence data includes non-WGS bulk submission types, that is, Expressed Sequence Tag (EST), Genome Survey Sequence (GSS), Patent and Transcriptome Shotgun Assembly (TSA). Whole Genome Shotgun (WGS) includes the number of sequence overlap contigs. Non-bulk data are the remainder.
Figure 2.
Cumulative base pairs in INSDC over time since 1980, broken down into selected data components. Data volume in base pairs of assembled sequence (whole genome shotgun methods and others) and raw next-generation-sequence data, excluding the Trace Archive (raw data from capillary sequencing platforms).
COLLABORATION FOR THE YEAR 2013
Members of the INSDC meet annually to discuss practical matters to maintain and develop the nucleotide sequence archives. Issues range from the addition of feature or qualifier elements to the feature tables present in the flat file report format in the traditional archive records to policy issues and strategies for dealing with the increasing sequence data to be archived. In 2012, the annual meeting was held at NCBI, Bethesda, MD USA, 11–13 June. At the meeting, we discussed and came to agreement on many issues. The outcomes of the meeting are summarized later in the text.
BIOSAMPLE DATABASE
With the emergence of high-performance nucleotide sequencing devices, the same biological sample can be analysed several times in the same or other project. For the convenience of data submitters, EBI (6) and NCBI (7) independently launched BioSample databases to store and provide sample information. The BioSample databases contain descriptions of biological source materials used in experimental assays. The purpose of the BioSample database is to provide unified storage and access to information about biological samples, which may have assay data stored in multiple databases. In 2012, DDBJ started to prepare to join this BioSample framework; in consequence, all INSDC members will collect and exchange sequence-related BioSample data as part of this new collaborative activity.
All organism names that are represented in the sequence data of INSDC are registered to the NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/taxonomy). Since 2009, the taxonomy database has considered terminating the assignment of strain-level taxonomy ID for microorganism genomes. However, taxonomy database agreed not to stop assigning strain-level taxonomy IDs for prokaryotic strains with sequenced genomes until at least 2013. The change to the current practice will not be made until BioSample has reached maturity and sample records representing these strains can be exchanged; hence, the change may not take place until later in 2013 or beyond.
TERMINATION OF MASS SEQUENCE FOR GENOME ANNOTATION SUBMISSION
Since 2004, INSDC has accepted the submission of Mass sequence for Genome Annotation (MGA) as one means of supporting large-scale of sequence data that provides information for annotation of genome assemblies/sequences. However, along with the popularization of new sequencing platforms, the MGA method has become out of date. Therefore, the INSDC decided to terminate accepting new submission of MGA data.
GENOME COLLECTION
Both submitters and users require INSDC to collect genome data with varied samples and study goals. Especially for bulk sequenced and re-sequenced genomes, INSDC requests that data providers to submit at least one set of assembled/annotated reference genome data, to submit raw reads to SRA for other genomes with associated Binary Alignment/Map (BAM), Variant Call Format (VCF) and General Feature Format (GFF) as ‘analysis’ objects; that is, without draft assembles of Whole Genome Shotgun (WGS) and scaffold Contig/Constructed (CON) data. Although in cases where genomes are sequenced/assembled to finished level, that is, possibly treated as a reference genome, INSDC should not label ‘complete genome’ in KEYWORDS section for genome data without feature annotation. The INSDC encourages submitters to annotate their sequences by providing tools and help documents describing minimal standards and requirements. NCBI introduced the Assembly database (http://www.ncbi.nlm.nih.gov/assembly) which has information about the structure of assembled genomes as represented in an AGP (A Golden Path) format file or as a collection of completely sequenced chromosomes. The INSDC members agreed to collaborate with this activity.
FUNDING
DDBJ by the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT); European Nucleotide Archive by the European Molecular Biology Laboratory, the Wellcome Trust, the FP7 Programme of the European Commission and the Biotechnology and Biological Sciences Research Council; NCBI by the Intramural Research Program of the National Institutes of Health; National Library of Medicine. Funding for open access charge: DDBJ management expense grant from MEXT, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Karsch-Mizrachi I, Cochrane G, Nakamura Y. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40:D33–D37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kodama Y, Shumway M, Leinonen L, on behalf of the International Nucleotide Sequence Database Collaboration The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodama Y, Mashima J, Kaminuma E, Gojobori T, Ogasawara O, Takagi T, Okubo K, Nakamura Y. The DNA Data Bank of Japan launches a new resource, the DDBJ Omics Archive of functional genomics experiments. Nucleic Acids Res. 2012;40:D38–D42. doi: 10.1093/nar/gkr994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amid C, Birney E, Bower L, Cerdeño-Tárraga A, Cheng Y, Cleland I, Faruque N, Gibson R, Goodgame N, Hunter C, et al. Major submissions tool developments at the European Nucleotide Archive. Nucleic Acids Res. 2012;40:D43–D47. doi: 10.1093/nar/gkr946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gostev M, Faulconbridge A, Brandizi M, Fernandez-Banet J, Sarkans U, Brazma A, Parkinson H. The BioSample Database (BioSD) at the European Bioinformatics Institute. Nucleic Acids Res. 2012;40:D64–D70. doi: 10.1093/nar/gkr937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]