Abstract
EuPathDB (http://eupathdb.org) resources include 11 databases supporting eukaryotic pathogen genomic and functional genomic data, isolate data and phylogenomics. EuPathDB resources are built using the same infrastructure and provide a sophisticated search strategy system enabling complex interrogations of underlying data. Recent advances in EuPathDB resources include the design and implementation of a new data loading workflow, a new database supporting Piroplasmida (i.e. Babesia and Theileria), the addition of large amounts of new data and data types and the incorporation of new analysis tools. New data include genome sequences and annotation, strand-specific RNA-seq data, splice junction predictions (based on RNA-seq), phosphoproteomic data, high-throughput phenotyping data, single nucleotide polymorphism data based on high-throughput sequencing (HTS) and expression quantitative trait loci data. New analysis tools enable users to search for DNA motifs and define genes based on their genomic colocation, view results from searches graphically (i.e. genes mapped to chromosomes or isolates displayed on a map) and analyze data from columns in result tables (word cloud and histogram summaries of column content). The manuscript herein describes updates to EuPathDB since the previous report published in NAR in 2010.
INTRODUCTION
The Eukaryotic Pathogen Database (EuPathDB: http://eupathdb.org) is one of the five NIAID/NIH-funded Bioinformatics Resource Centers (BRCs) supporting infectious disease pathogens and invertebrate vectors of human disease (1–5). BRC resources provide free online access to functional genomic data with tools that enable integrated data interrogation (6). Additional information regarding the BRC program is available on NIAID websites (http://www.niaid.nih.gov/labsandresources/resources/dmid/brc/Pages/default.aspx) and the BRC portal site (http://pathogenportal.org).
EuPathDB is specifically tasked with providing support to research communities investigating eukaryotic pathogens, in particular (but not limited to) categories A–C priority and (re)-emerging pathogens. In addition, collaborative efforts between EuPathDB and GeneDB (7) with funding from The Bill and Melinda Gates Foundation and The Wellcome Trust made it possible to develop a kinetoplastid resource (8). Currently, EuPathDB includes 11 component sites listed with their web addresses in Table 1 (1,8–13). All databases incorporate the Strategies WDK, a unique graphical search tool that enables users to perform complex combinatorial queries (14). This system has been used by other genomic resources including, FungiDB (http://fungidb.org) (15), SchistoDB (http://schistodb.net) (16), TBDB (http://www.tbdb.org/wdk/) (17) and BetaCell (http://www.betacell.org) (18).
Table 1.
This table lists EuPathDB resources, their web addresses and the included organisms
| Database | Web address | Supported organisms |
|---|---|---|
| EuPathDB | http://eupathdb.org | All EuPathDB organisms listed below |
| AmoebaDB | http://amoebadb.org | Entamoeba histolytica, E. dispar, E. invadens, E. moshkovskii |
| CryptoDB | http://cryptodb.org | Cryptosporidium parvum, C. hominis, C. muris |
| GiardiaDB | http://giardiadb.org | Giardia lamblia assemblages A, B and E |
| MicrosporidiaDB | http://microsporidiadb.org | Edhazardia aedis, Encephalitozoon cuniculi, E. hellem, E. intestinalis, Enterocytozoon bieneusi, Hamiltosporidium tvaerminnensis, Nematocida parisii, Nosema ceranae, Vavraia culicis |
| PiroplasmaDB | http://piroplasmadb.org | Babesia bovis, Theileria annulata, T. parva |
| PlasmoDB | http://plasmodb.org | Plasmodium berghei, P. chabaudi, P. falciparum, P. gallinaceum, P. knowlesi, P. reichenowi, P. vivax, P. yoelii |
| ToxoDB | http://toxodb.org | Toxoplasma gondii, Eimeria tenella, Gregarina niphandrodes, Neospora caninum |
| TrichDB | http://trichdb.org | Trichomonas vaginalis |
| TriTrypDB | http://tritrypdb.org | Trypanosoma brucei, T. congolense, T. cruzi, T. vivax, Leishmania major, L. infantum, L. braziliensis, L. Mexicana, L. panamensis, L. tarentolae, Endotrypanum monterogeii |
| OrthoMCL | http://orthomcl.org | Includes proteins from over 150 organisms across bacteria, archaea and eukarya. |
WHAT IS NEW IN EuPathDB
Over the past 2 years, EuPathDB has made advances in its repertoire of databases, data content, analysis and visualization tools and its infrastructure.
New databases
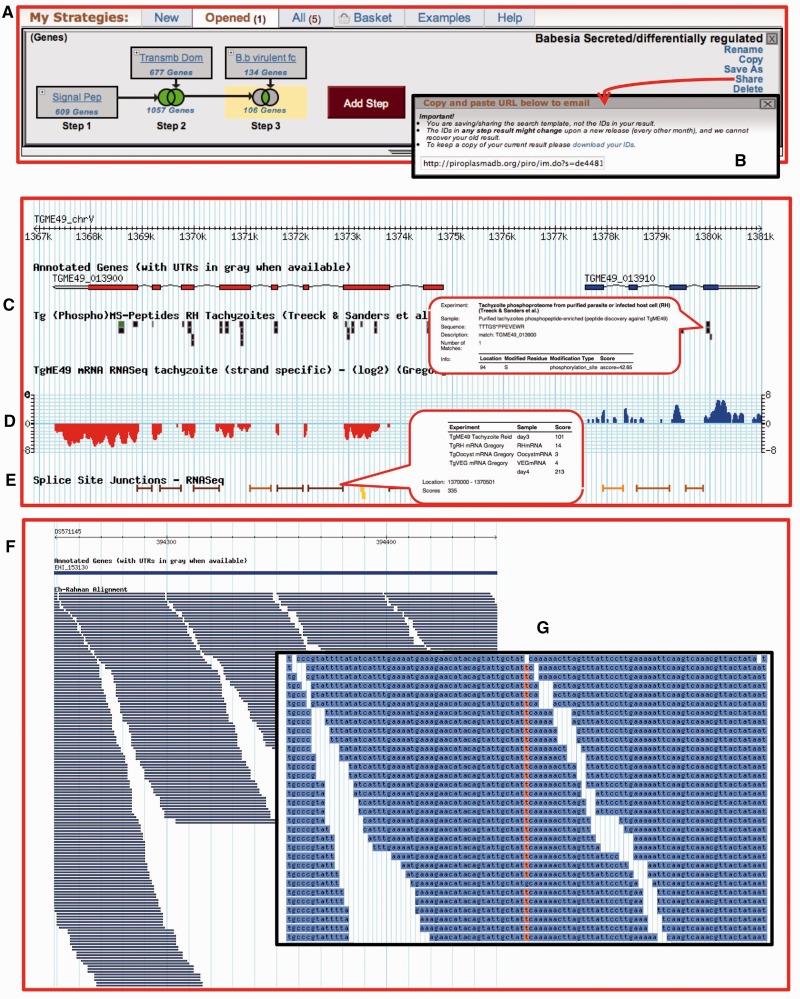
The latest addition to the EuPathDB family of databases is PiroplasmaDB (http://piroplasmadb.org), which supports Babesia and Theileria parasites. The look and feel of PiroplasmaDB is identical to other EuPathDB resources. Searches in this database are conducted using the search strategy system (14), which involves the sequential addition of searches using set operations to produce a refined list of results (11). Figure 1A depicts a search strategy in PiroplasmaDB that defines a list of genes predicted to contain signal peptides, transmembrane domains or both, and are differentially regulated between a virulent and an attenuated strain of Babesia bovis (19). To facilitate collaborative efforts, search strategies may be shared using a uniquely generated URL (Figure 1B). For example, the search strategy displayed in Figure 1A may be accessed using the following address: http://piroplasmadb.org/piro/im.do?s=de44813e1905d647.
Figure 1.
Screen shots of a search strategy in PiroplasmaDB and GBrowse representing HTS (C–E from ToxoDB and F and G from AmoebaDB) (A) A three-step search strategy combining genes with predicted signal peptides, transmembrane domains and microarray expression data. (B) Search strategies may be saved and shared with others using a uniquely generated URL. (C) Peptides from mass spec experiments are mapped to genes and displayed graphically. Mousing over the graphics provides additional information, such as the peptide sequence and any posttranslational modifications. In this image, peptides are from a phophoproteomic experiment. (D) A track representing strand-specific RNA-seq data. Blue indicates reads mapping to the forward strand, whereas red represents those mapping to the reverse strand. (E) Unified splice junction track representing intron-spanning RNA-seq reads from all experiments in the database. (F) A 2 kb region with alignment of DNA sequencing reads to the genome. (G) Zooming in to 100 bp displays the actual sequence allowing data inspection. Highlighted nucleotides represent SNPs.
ReFlow workflow system
The EuPathDB data builds are complex because the project includes 11 different websites, each with its own underlying database. In each bi-monthly release cycle, some of these databases are completely rebuilt (when there are major changes to multiple genomes). The rest may receive incremental updates to add high-value data sets, such as newly sequenced and annotated genomes or new functional experiments or to revise existing ones. In both cases, the build is controlled entirely by workflows using the ReFlow workflow system developed in-house. The workflows are dependency graphs specifying every step of creating the integrated database, from data acquisition, through analysis on a compute cluster, to cross-referencing and finally loading. As an example, PlasmoDB’s workflow has approximately 5000 distinct steps, which analyze and load data from approximately 250 data sets. ReFlow is uniquely suited to building genomic databases as it supports running ‘in reverse’ to remove outdated data. ReFlow is used during each build cycle to revise outdated data sets, to recompute cross-genome analyses when we add new genomes and to redo data that our QA process has identified as having a bug.
New data content
The data content in EuPathDB has increased both in quantity and type. An updated data content table is available at the following URL: http://eupathdb.org/eupathdb/showXmlDataContent.do?name=XmlQuestions.GenomeDataType
Genome sequence and annotation
The number of available sequenced and annotated genomes has increased dramatically owing in large part to the presence of a number of sequencing ‘white papers’ specifically tasked with sequencing eukaryotic pathogens (i.e. The Broad Institute—Plasmodium and Microsporidia; the J. Craig Venter Institute—Toxoplasma and Entamoeba; and the Genome Institute at Washington University—Kinetoplastida). Additional whole-genome sequencing data are provided by the parasite genomics section of the Sanger Institute and individual research laboratories. EuPathDB incorporates both annotated and unannotated genomes providing searches based on the provided data (i.e. annotation, BLAST analysis, sequence retrieval and download, etc.) and based on various analyses performed in-house [i.e. InterPro scan (20), open reading frame prediction, BLAT against the NCBI, Genome Ontology searches, searches against available functional data, etc.].
New data types include
Phosphoproteomic data
Mass spectrometry-based data representing peptides with phosphorylated amino acids have been incorporated allowing users to search for genes with modified peptides and graphically visualize modified peptides. Figure 1C shows a Genome Browser (GBrowse) (21) view from ToxoDB showing phospho-peptides mapped against genes (22). Mousing over the peptide glyphs reveals information regarding the peptide amino acid sequence, modified amino acid and genomic location.
Strand-specific RNA sequence (RNA-seq) data
Data from such experiments are represented in GBrowse as histograms of depth of read coverage. Reads aligning to the forward strand are in blue and those aligning to the reverse strand are in red (Figure 1D). Currently, strand-specific RNA-seq data are available in PlasmoDB (Newbold and Berriman groups, unpublished data) and ToxoDB (Boothroyd and Gregory groups, unpublished data).
Splice junction predictions (based on RNA-seq)
Intron-spanning RNA-seq reads are aligned to the genome using the RNA-seq unified mapper (23) (Figure 1E). Intron-spanning reads from individual experiments or from all available experiments combined may be visualized. Mousing over intron spans reveals experimental information and the number of reads that support the span enabling users to evaluate the confidence of the intron and identify genes that show evidence for alternative RNA processing.
Single nucleotide polymorphism data based on high-throughput sequencing
Single nucleotide polymorphisms (SNPs) based on high-throughput sequencing (HTS) data are determined by aligning reads to the reference genome using Bowtie (24), post-processing with SamTools (25) and GATK (26) and ultimately called using VarScan (27). Genes can be identified based on their SNP characteristics and parameters, such as allele frequency (based on percent allele-matched reads), P-value and depth of coverage supporting a SNP may be tweaked. Read pileup data are available in GBrowse, including the ability to view actual aligned sequence reads (Figure 1F and G) to further assess the quality of individual SNP calls.
Expression quantitative trait loci data
Genes may be identified based on their association to genome-wide expression-level polymorphisms from a genetic cross between phenotypically distinct parasite clones of Plasmodium falciparum (HB3 and Dd2) (28). This data may be searched and visualized in multiple ways.
Genes may be identified based on their association to genomic segments, expression profile similarity or similarity of genetic association. Genomic segments can be identified based on their association to genes. Regions/spans that are associated by expression quantitative trait loci data (eQTL) are displayed in a table on gene pages and both microsatellites and haplotype blocks are available as tracks in GBrowse.
High-throughput phenotyping data
Essential Trypanosoma brucei genes can be identified based on the decreased sequence read coverage generated from sequencing the population of expression library cassettes in a genome-wide RNAi-based screen (29). The high-throughput phenotyping search is located in the ‘Putative Function’ section under the heading ‘Identify Genes by’ on the TriTrypDB home page (8). A sample strategy that searches this data for genes that are likely essential in all stages or time points examined can be accessed here: http://tritrypdb.org/tritrypdb/im.do?s=0e54e90e623cbbc2
Graphs and tables representing the expression and percentile values for individual genes are available in the ‘Phenotype’ section of gene pages, and GBrowse tracks of coverage plots for each sample from this experiment are available.
New Tools
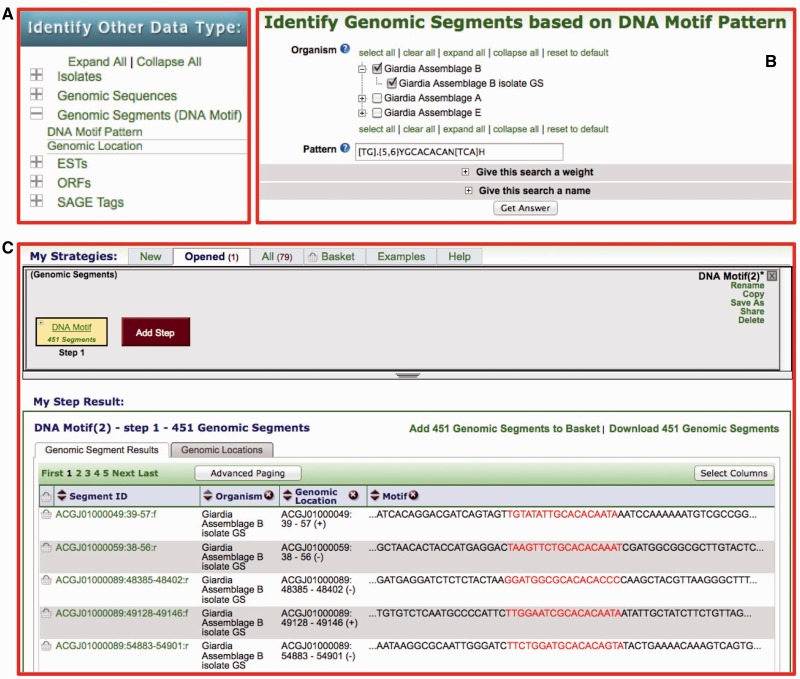
Genomic segment tool
DNA segments may be defined based on their genomic location or their nucleotide sequence (DNA motif pattern) (Figure 2A). This search dynamically generates segment records allowing the incorporation of results into a search strategy (see genomic colocation, below). This new search is available under ‘Identify Other Data Types’; click on ‘Genomic Segments (DNA motif)’ then select either ‘DNA motif pattern’ or ‘Genomic location’ (Figure 2A). Figure 2B shows the DNA motif pattern search page, which allows selection of target organisms to search (example shown from GiardiaDB) and an input window for the DNA motif pattern (simple text or a regular expression may be used). Results of a DNA motif pattern search are returned as a step in a strategy and the motif records are displayed including the identified motif (Figure 2C).
Figure 2.
Screen shot from GiardiaDB depicting a genomic segment search. (A) Genomic segment searches (i.e. DNA motif pattern) are available on the home page. (B) DNA motifs may be entered as a standard string of characters or using a regular expression as depicted. (C) DNA segment records are generated dynamically and results are displayed in a search strategy with results represented in a dynamic table below the strategy.
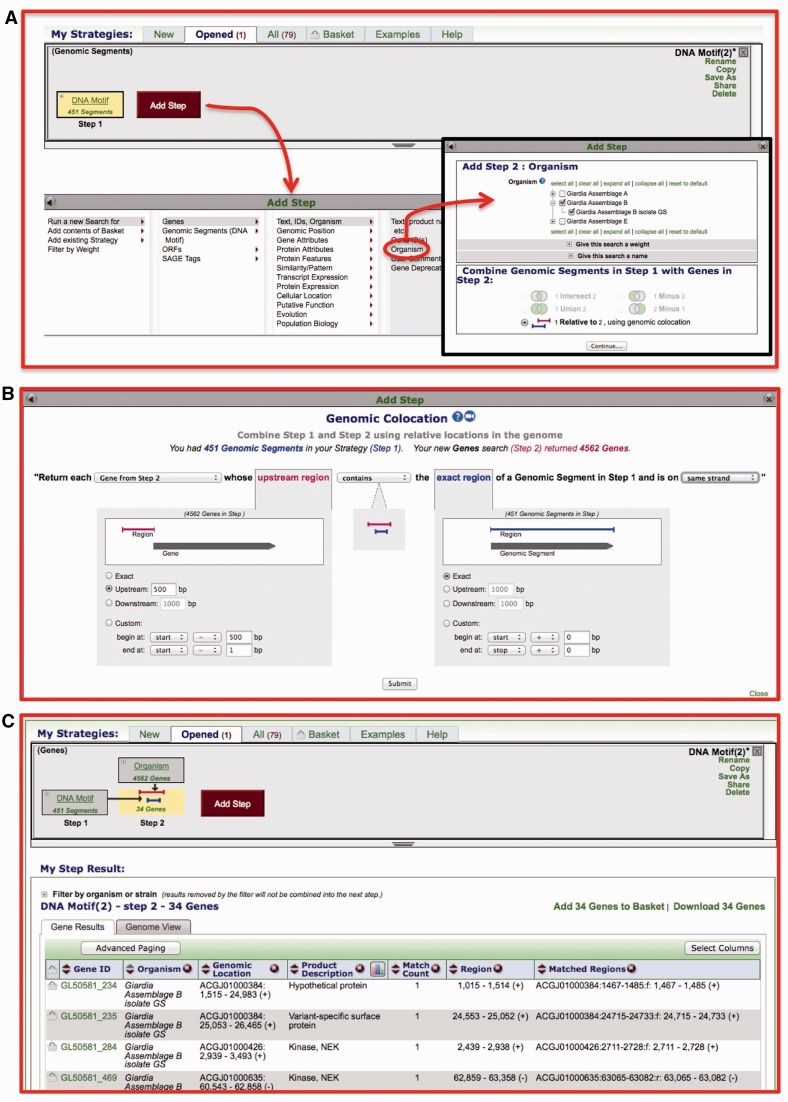
Genomic colocation tool
This tool enables searches based on a user-defined relationship between entities with defined genomic coordinates (i.e. genes, SNPs, DNA motifs, etc.). For example, one may be interested in identifying all genes that have a SNP or a DNA motif located within 500-nt upstream of the 5′-end. Figure 4 illustrates the steps taken to find all genes that have a DNA motif defined in Figure 3C located within 500-nt upstream of the 5′-end. After running a DNA pattern search, a step is added to define all genes in the organism of interest (Figure 3A). Since the steps in this strategy include different result types (DNA motifs and genes), the only option available for combining the results is the genomic colocation option (Figure 3A). The next step is to define which results to retrieve based on the user-defined colocation relationship (Figure 3B). The customizable colocation popup provides a dynamic logic statement that is updated based on the chosen parameters (Figure 3B). Once the parameters are set, the logic statement in this example is ‘Return each gene from step 2 whose upstream region contains the exact region of a Genomic Segment from step 1 and is on the same strand’. Clicking on ‘Get Answer’ returns all genes that meet the colocation criteria (results include in addition to gene IDs, the number and location of matches (Figure 3C).
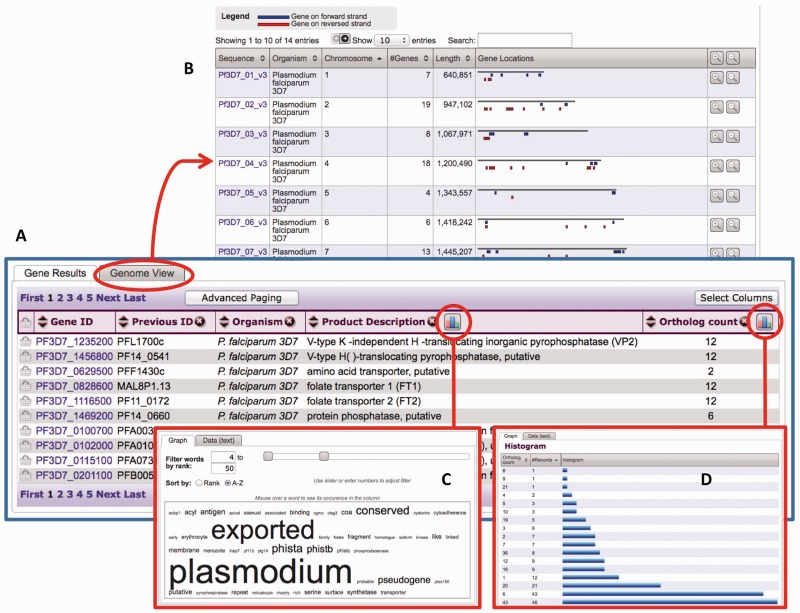
Figure 4.
Screen shots from PlasmoDB showing in (A) a typical result list from a search strategy, (B) an alternative graphical representation of genes on chromosomes, (C) a word cloud generated by clicking on the column analysis icon for the product description column and (D) a histogram generated by clicking on the column analysis icon for the ortholog count column.
Figure 3.
Screen shots depicting the genomic colocation query in EuPathDB resources. In this example from GiardiaDB, genes that have a DNA motif located within 500-nt upstream are identified. (A) To identify genes in relation to DNA motifs, a step searching for genes based on the organism of interest is added to the strategy. The genomic colocation option is selected by default when combing different record types, such as DNA motifs and genes. (B) The customizable colocation popup provides a dynamic logic statement that is updated based on the chosen parameters. (C) Results of colocation query. Top of the panel shows the search strategy and the bottom portion includes the results with columns for gene IDs, number of matched motifs in the defined region and match genomic coordinates.
Alternative views of search results
Search results are typically visualized as a list of results in a table with customizable columns (Figure 4A) (1). A new feature provides tabs that enable users to choose alternative data views. For example, in gene results pages, users can choose a graphical visualization of their genes mapped on the genome (Figure 4B) to determine, if there is bias in the genomic distribution. A user may zoom in on individual chromosomes and click on the gene graphic to visit the gene page or a GBrowse view. For isolate results, users can select a Google map view to visualize the geographic distribution of the isolates. Clicking on the pins pops up, specific information with the option to retrieve isolate results from that country.
Column analysis
This tool enables users to analyze data within columns of the results table after running a search. To access this feature, run any search that returns a list of results, then click on the icon next to the column name (Figure 4A). Currently, this tool offers two analyses: word clouds for columns containing text (Figure 4C) and histograms for columns containing numbers (Figure 4D). Further analyses, including enrichment analysis for GO terms, EC numbers and pathways, will be implemented in the near future.
Updated Genome Browser
The GMOD Genome Browser has been updated to version 2.48. The update provides several new GBrowse features to EuPathDB users, including the ability to upload BAM files in the custom tracks section allowing private display of HTS data in the context of other available data tracks. Additional features available in GBrowse may be accessed at the following URL: http://gmod.org/wiki/GBrowse_2.0_HOWTO
Future directions
EuPathDB resources will continue to expand both in data content and type, and in functionality. Development projects that are currently underway include:
integration of OrthoMCL into the strategiesWDK: this would facilitate better integration of data from OrthoMCL with the rest of EuPathDB and would promote integrated evolution-based queries using search strategies;
incorporation of mass spectrometric metabolomic data allowing queries for changes in the metabolome of parasites in response to developmental or environmental changes;
incorporation of parasite host response data enabling users to ask questions regarding changes in host cells (i.e. RNA-seq, microarray, proteomics, etc.) in response to infection by eukaryotic parasites;
enabling direct data export from EuPathDB, a Galaxy server (30). This would allow users to perform custom analysis with data obtained from EuPathDB and their own uploaded data. Examples of this include analysis of RNA-seq results, SNP analysis and phylogenetic tree reconstruction; and
enabling GBrowse login to allow users to store their custom tracks and GBrowse preferences in their EuPathDB user profile.
FUNDING
National Institute of Allergy and Infectious Diseases (EuPathDB); National Institutes of Health, Department of Health and Human Services [Contract No. HHSN272200900038C to D.S.R., C.J.S. and J.C.K.]; Bill and Melinda Gates Foundation, The Wellcome Trust [WT085822MA, The TriTrypDB component of EuPathDB]. Funding for open access charge: NIH Contract No. [HHSN272200900038C].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank members of the EuPathDB research communities for their willingness to share genomic-scale data sets, often prior to publication and for numerous comments and suggestions from our scientific advisors and the scientific community at large, which have helped to improve the functionality of EuPathDB resources. We also thank past and present staff associated with the EuPathDB BRC project, and our research laboratory colleagues whose contributions have facilitated the creation and maintenance of this database resource.
REFERENCES
- 1.Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Squires RB, Noronha J, Hunt V, García Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, et al. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respi Viruses. 2012;6:404–416. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2011;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DST, Koscielny G, Louis C, Maccallum RM, Redmond SN, et al. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40:D729–D734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, Driscoll T, Hix D, Mane SP, Mao C, et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 2011;79:4286–4298. doi: 10.1128/IAI.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene JM, Collins F, Lefkowitz EJ, Roos D, Scheuermann RH, Sobral B, Stevens R, White O, Di Francesco V. National Institute of Allergy and Infectious Diseases Bioinformatics Resource Centers: New Assets for Pathogen Informatics. Infect Immun. 2007;75:3212–3219. doi: 10.1128/IAI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logan-Klumpler FJ, De Silva N, Boehme U, Rogers MB, Velarde G, McQuillan JA, Carver T, Aslett M, Olsen C, Subramanian S, et al. GeneDB–an annotation database for pathogens. Nucleic Acids Res. 2012;40:D98–D108. doi: 10.1093/nar/gkr1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2009;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, Rhodes P, Wang S, He C-Z, Su Y, et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34:D419–D422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aurrecoechea C, Barreto A, Brestelli J, Brunk BP, Caler EV, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al. AmoebaDB and MicrosporidiaDB: functional genomic resources for Amoebozoa and Microsporidia species. Nucleic Acids Res. 2011;39:D612–D619. doi: 10.1093/nar/gkq1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer S, Aurrecoechea C, Brunk BP, Gao X, Harb OS, Kraemer ET, Pennington C, Treatman C, Kissinger JC, Roos DS, et al. The Strategies WDK: a graphical search interface and web development kit for functional genomics databases. Database. 2011;2011:bar027. doi: 10.1093/database/bar027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, et al. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2012;40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerlotini A, Heiges M, Wang H, Moraes RLV, Dominitini AJ, Ruiz JC, Kissinger JC, Oliveira G. SchistoDB: a Schistosoma mansoni genome resource. Nucleic Acids Res. 2009;37:D579–D582. doi: 10.1093/nar/gkn681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galagan JE, Sisk P, Stolte C, Weiner B, Koehrsen M, Wymore F, Reddy TBK, Zucker JD, Engels R, Gellesch M, et al. TB database 2010: overview and update. Tuberculosis. 2010;90:225–235. doi: 10.1016/j.tube.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Mazzarelli JM, Brestelli J, Gorski RK, Liu J, Manduchi E, Pinney DF, Schug J, White P, Kaestner KH, Stoeckert CJ. EPConDB: a web resource for gene expression related to pancreatic development, beta-cell function and diabetes. Nucleic Acids Res. 2007;35:D751–D755. doi: 10.1093/nar/gkl748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau AO, Kalyanaraman A, Echaide I, Palmer GH, Bock R, Pedroni MJ, Rameshkumar M, Ferreira MB, Fletcher TI, McElwain TF. Attenuation of virulence in an apicomplexan hemoparasite results in reduced genome diversity at the population level. BMC Genomics. 2011;12:410. doi: 10.1186/1471-2164-12-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 21.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe. 2011;10:410–419. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11, Unit 11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, Wuchty S, Rathod PK, Ferdig MT. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol. 2008;6:e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alsford S, Eckert S, Baker N, Glover L, Sanchez-Flores A, Leung KF, Turner DJ, Field MC, Berriman M, Horn D. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goecks J, Nekrutenko A, Taylor J Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]