Abstract
Genome duplication (GD) has permanently shaped the architecture and function of many higher eukaryotic genomes. The angiosperms (flowering plants) are outstanding models in which to elucidate consequences of GD for higher eukaryotes, owing to their propensity for chromosomal duplication or even triplication in a few cases. Duplicated genome structures often require both intra- and inter-genome alignments to unravel their evolutionary history, also providing the means to deduce both obvious and otherwise-cryptic orthology, paralogy and other relationships among genes. The burgeoning sets of angiosperm genome sequences provide the foundation for a host of investigations into the functional and evolutionary consequences of gene and GD. To provide genome alignments from a single resource based on uniform standards that have been validated by empirical studies, we built the Plant Genome Duplication Database (PGDD; freely available at http://chibba.agtec.uga.edu/duplication/), a web service providing synteny information in terms of colinearity between chromosomes. At present, PGDD contains data for 26 plants including bryophytes and chlorophyta, as well as angiosperms with draft genome sequences. In addition to the inclusion of new genomes as they become available, we are preparing new functions to enhance PGDD.
INTRODUCTION
Most higher organisms pass through different ploidy levels at different stages of development (1,2) and continuously produce aberrant unreduced gametes at low rates. However, the extreme rarity of genome duplications (GDs) in the evolutionary history of extant lineages, occurring only once in many (sometimes hundreds of) millions of years, shows that the vast majority of GD events quickly go extinct. For the rare survivors, classical views suggest that GD is potentially advantageous as a primary source of genes with new (3,4) or modified functions (5).
The angiosperms (flowering plants) are an outstanding model in which to elucidate consequences of GD in higher eukaryotes. Gene-order conservation in vertebrates is evident after hundreds of millions of years of divergence (6,7). However, the two major branches of the angiosperms (eudicots and monocots), estimated to have diverged 125–140 MY (8) to 170–235 MYA (9) show much more rapid structural evolution, owing largely to their propensity for chromosomal duplication and subsequent gene loss (10), fragmenting ancestral linkage arrangements across multiple chromosomes (11–13). All angiosperm genomes published to date have shown evidence of paleopolyploidy (14). Although new data from yeast (15–17) and Paramecium (18) are shedding valuable light on consequences of GD in microbes, these consequences are expected to be very different in organisms with small effective population sizes such as angiosperms, mammals and other higher eukaryotes (19,20). For example, neofunctionalization is much more likely to occur in large populations, which contain more targets for mutations conferring new beneficial function. In contrast, subfunctionalization is improbable in large populations, as a partially subfunctionalized allele (the first step in the process) is more likely to be silenced by secondary mutations before reaching fixation by drift (19).
A host of investigations into the functional and evolutionary consequences of gene and GD may be empowered by genome alignments from a single resource based on uniform standards that have been validated by empirical studies. Algorithms commonly used in vertebrate genome alignments focus on identifying orthologous regions, as GDs are rare, and ancient and paralogous regions are often so diverged as to be unrecognizable. However, to reveal the consequences of the more recent and more frequent GDs in angiosperms and other taxa, identifying paralogous regions is of central importance, necessitating the use of multiple alignments, both within and among genomes. To tackle such problems, we implemented a multiple gene-order alignment tool MCScan, which reflects better the true relationships among angiosperm genomes, in which GDs are frequently superimposed on speciations (21). Further, to empower comparative and functional studies across (and potentially beyond) the burgeoning set of plant genome sequences available, we built the Plant Genome Duplication Database (PGDD), a web service providing synteny information in terms of gene colinearity, both within and between genomes.
Besides PGDD, comparative genomic data are available from some public databases such as CoGe (22), Phytozome (23), GreenPhylDB (24) and PLAZA (25). CoGe (22) provides comparative data across all species in any state of assembly by computation on the fly while this allows greater flexibility on the user-end, non-specialists who are searching for a well-curated resource may find it cumbersome to use. In green plant, Phytozome (23) and GreenPhylDB (24) provide well-controlled micro-synteny and gene family evolution data, but macro-synteny data are not supported by the databases. PLAZA (25) provides fine macro-synteny data in plants, as well as micro-synteny and gene family data such as PGDD. However, there are some differences between PGDD and PLAZA in colinearity data because to identify colinear gene pairs, PLAZA adopted i-ADHoRe (26) of which power and precision differ from MCScan (27) used in PGDD.
For the past 5 years, PGDD has provided data about syntenic relationships based on colinear blocks between plants and contributed to much research such as evolution of gene families (28–34), annotations (35–38) and polyploidy events (39–43). PGDD also provides an easily linked data web resource to be readily integrated to other external informatics portal, including TAIR (44), Legume Information System (45) and PopGenIE (46). In the past year alone, we have developed a new pipeline to promptly merge new genome data into the database and nearly tripled the number of genomes archived. At present, PGDD contains data for 26 plants including bryophytes and chlorophytes, as well as angiosperms (Table 1).
Table 1.
List of 26 plants currently served by PGDD
| Species name | Common name | Release version | Gene number |
|---|---|---|---|
| Arabidopsis lyrata | Lyrate rockcress | Version 1.0 (April 2011) | 32 670 |
| Arabidopsis thaliana | Arabidopsis | TAIR 9.0 (June 2009) | 27 379 |
| Brachypodium distachyon | Purple false brome | Phytozome v6.0 | 32 255 |
| Brassica rapa | Chinese cabbage | Version 1.1 | 22 285 |
| Cajanus cajan | Pigeonpea | November 2011 | 48 680 |
| Carica papaya | Papaya | December 2007 | 25 536 |
| Chlamydomonas reinhardtii | Green algae | Version 4.2 | 16 036 |
| Cucumis sativus | Cucumber | Phytozome v6.0 | 21 491 |
| Fragaria vesca | Strawberry | December 2010 | 34 809 |
| Glycine max | Soybean | Release 1 (December 2008) | 66 153 |
| Lotus japonicus | Lotus | Release 2.5 | 42 399 |
| Malus × domestica | Apple | August 2010 | 57 386 |
| Medicago truncatula | Barrel medic | Mt 3.5.1 (December 2010) | 45 108 |
| Musa acuminata | Banana | July 2012 | 36 542 |
| Oryza sativa | Rice | RAP 2.0 (November 2007) | 30 192 |
| Physcomitrella patens | Moss | Version 1.6 (January 2008) | 32 272 |
| Prunus persicaa | Peach | Version 1.0 | 27 864 |
| Populus trichocarpa | Western poplar | Phytozome 2.0 (February 2010) | 45 778 |
| Ricinus communis | Castor bean | Release 0.1 (May 2008) | 38 613 |
| Sorghum bicolor | Sorghum | Sbi 1.4 (December 2007) | 34 496 |
| Solanum lycopersicum | Tomato | Release 2.3 | 34 727 |
| Solanum tuberosum | Potato | Version 3.4 | 39 031 |
| Selaginella moellendorffii | Spikemoss | Version 1.0 (December 2007) | 22 273 |
| Theobroma cacao | Cacao | Release 0.9 (September 2010) | 28 798 |
| Vitis vinifera | Grape vine | Genoscope (August 2007) | 26 346 |
| Zea mays | Maize | Release 5a (November 2010) | 32 540 |
aUnpublished genome data temporarily restricted for downloading (in accordance with the understandings in the Fort Lauderdale meeting and NHGRI policy statement).
DATABASE CONSTRUCTION
Data source
At present, the PGDD contains colinear block information within and between the genomes of 26 plants (Table 1), most recently updated to include the banana genome sequence published in August 2012 (47). Among them, 16 genomes were downloaded from the homepages of the institute that led the sequencing of the genome such as RAP-DB (Rice annotation project database; http://rapdb.dna.affrc.go.jp/) and BRAD (The Brassica database; http://brassicadb.org/brad/). Data for the remaining 10 plants, mostly sequenced by the US Department of Energy Joint Genome Institute, were downloaded from the Phytozome database (23). To build PGDD data, three types of file are used: coding DNA sequences file, protein sequences file and general feature format (GFF) file containing annotation data of the sequences in chromosomes.
Pipeline to analyse and add the new genome data
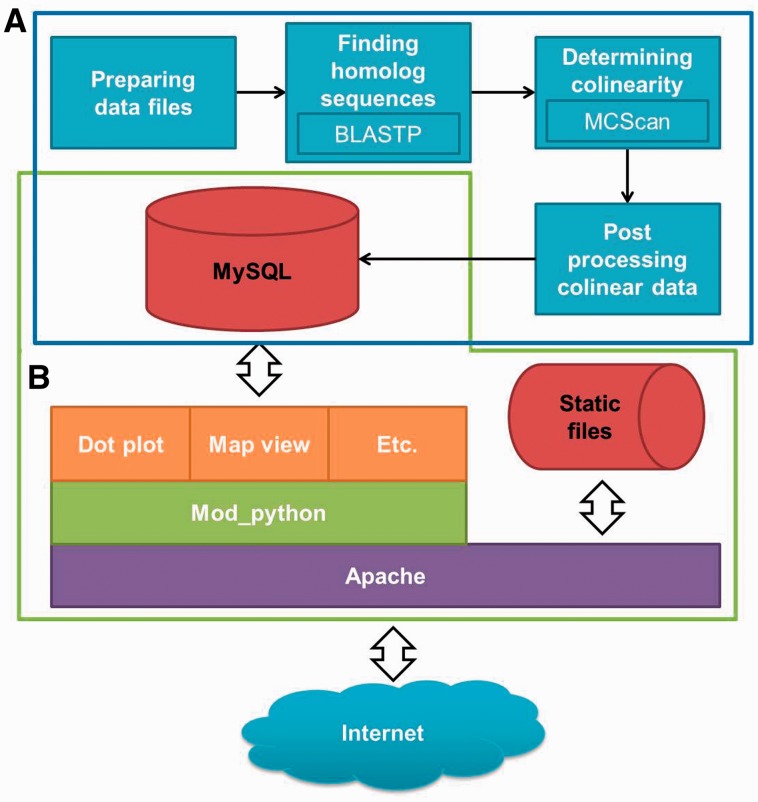
There are four major steps to add a new genome into PGDD (Figure 1A) in a pipeline consisting of 18 scripts. In the first step, scripts determine basic information such as the length of chromosomes and prepare data files. For example, one script extracts information of genes from a GFF file and makes a browser extensible data (BED) file to simply determine gene loci. Then, similar protein pairs are determined between two plants by BLASTP with 1e–5 e-value cut-off in the second step. The colinear blocks between plants are determined in the third step. With the BED file containing loci information and the file containing pairs of similar proteins created in the second step, colinear blocks between the plants are determined by MCScan (27). In the post-processing step, additional data are calculated and determined. For example, Ks values between pairs of ortholog/paralog genes are determined by Clustal W (48), PAL2NAL (49) and yn00 program of the PAML package (50) in this step. Additionally, text files containing all information about colinear blocks are created. Finally, all new blocks information included in the text files is imported into MySQL, and parameters and contents in PGDD web pages are modified for the new data by scripts.
Figure 1.
Diagram of current PGDD server. (A) Diagram of pipeline to update PGDD with new genome data (in blue box). The boxes in the diagram represent four major steps of the pipeline, consisting of 18 in-house scripts. Insets in some boxes contain the name of a major program in the process. (B) Layers diagram of PGDD structure (in green polygon).
Implementation of database
All scripts such as components in the pipeline to add new genome data were developed using Python programming language (http://www.python.org) and in Bash (http://www.gnu.org/software/bash/). MCScan was developed using the C++ programming language that has good run-time performance because of the huge number of calculations required to determine colinear blocks. Python was also used as a server-side web programming language. Thus, the developed server-side python scripts are running by mod_python (http://www.modpython.org/) on the Apache HTTP server (http://httpd.apache.org/) environment (Figure 1B). To draw plots in Python, matplotlib (http://matplotlib.sourceforge.net/) was mainly used. As a client-side web programming language, we adopted JavaScript because most web browsers support this language well. However, to overcome problems caused by differences between browsers, jQuery (http://www.jquery.com) was used as the JavaScript library.
All colinear blocks and related data provide by PGDD are stored in a MySQL database (http://www.mysql.com). There are three major tables, block, locus and chromosome, in the database. The block table contains lists of gene pairs with additional information such as colinear block number and Ks value, whereas the locus table contains information about each locus such as functional description determined by BLAST against Non-redundant GenBank DB and positions of loci in a chromosome. Information for each chromosome, such as number of genes in a chromosome, is stored in the chromosomes table. Besides MySQL, we maintain up-to-date protein sequences stored as BLAST database file to use in BLAST search function of PGDD.
WEB INTERFACES AND USAGE
The home page and major functions provided by PGDD
At the home page, PGDD shows a table containing information about all plants in the current version, including the name of a plant, version of genome used, number of genes, original URL to download the data and primary citation for the genome (Figure 2A). Additionally, the table provides related web links, such as taxonomy information at NCBI, so that users can easily get related information for each plant.
Figure 2.
Homepage and examples of three major functions of PGDD. (A) The homepage of PGDD and functions supported by the database. (B) Web page of Dot plot function and a plot applying a Ks filter of 0.4–0.7 between rice and sorghum as an example, representing colinear blocks between the plants. (C) Example of Locus-search result for AT1G25460 loci in Arabidopsis. A blue line in alignment image represents same orientations of paired genes, whereas the red line represents opposite orientations of the genes. (D) Example of Map-view function. The grey vertical bars represent chromosomes, and green arrows on the bars represent the position of locus, which are similar to input sequences. The detailed loci information page, the inset, shows protein and nucleotide sequences of gene in the loci and a description of the gene.
There are three major functions to show gene colinearity; Dot-plot, Locus-search and Map-view. These three functions provide means to visualize macro-synteny, micro-synteny and gene family evolution, respectively, which is often the most commonly needed information in comparative genomics research. The main menu to select a web page corresponding to each function is in the right of the table. In addition to the major functions, in the download page, a file containing colinear block information within a plant or between any 2 of the 26 plants can be downloaded.
Dot-plot module to show overall view of colinear blocks
Dot plots are used to show colinear blocks between two plant genomes in macro-scale as a two-dimensional image, so researchers can see the overall view of all blocks. For example, the dot plot in Figure 2B shows overall colinear blocks between rice and sorghum including both the orthologous regions and matching regions derived from a shared pan-cereal duplication event (ρ) (51). Each point represents a matched gene pair. Interpretation of a dot plot is not always straightforward because diverse events in evolutionary history are overlaid onto the same plot. Thus, many options are provided to modify the plot through filtering subsets of gene pairs, e.g. to show only a narrow range of synonymous substitutions (Ks values) of gene pairs as a proxy to separate the gene pairs by age. Using rice-sorghum as an example, applying a Ks filter of 0.4–0.7 renders signal from the orthologous gene pairs more prominent on the dot plot. Additionally, an enlarged dot plot between specific chromosomes is available by clicking each small box for each chromosome in the genome-wide plot. Besides the dot plot, users can see the list of gene pairs in colinear blocks by clicking on the segment in the enlarged plot. With the list containing both name and inferred functions of the genes, users can compare colinear blocks with single-gene resolution.
Locus-search module to search locus in the database by name
There have been many gene-level studies such as comparing a few genes included in colinear blocks (28–38). Locus search is a function to find a colinear block containing a specific locus (Figure 2C) and to show fine structure of the colinear block. By typing the locus name in textbox and clicking the ‘submit’ button, a user can search colinear blocks containing the locus. Locus-search results can be divided into two parts: an alignment image and a list of genes in the image. In the alignment image, PGDD shows genes in colinear blocks, so users can easily determine gene-level changes such as insertion and deletion of genes. The list of genes below the each alignment image shows not only the inferred function of each gene but also the Ks and Ka values of the gene pair. Thus, the user easily determines evolutionary distance and possible changes in function between genes.
Map-view module to map locus on a chromosome
In many cases, a researcher seeks information about a locus just with a nucleotide or a protein sequence, without additional information such as locus name. A typical BLAST search returns a list of likely homologues in the target genomes but lack the global view of how the hits are distributed. To support such cases, PGDD provides Map View function. In the corresponding web page, users can search for a locus in PGDD by similarity with a nucleotide or protein sequence (Figure 2D). The page contains a text box to type or paste a sequence, buttons to choose a BLAST program depending on the sequence type and a text box to set e-value cut-off. The search result can be divided in two parts: list of locus names that are similar with user input sequence and image to show the positions of the locus in chromosomes. In the image, each grey vertical bar represents each chromosome, and each green arrow shows the positions of loci, which are similar to the sequence. The user can see detailed information for the locus and colinear blocks alignment image by clicking a blue locus name in the list of locus names above the image. In the detailed information page, the user can get protein and nucleotide sequences of genes in the locus, as well as descriptions of the genes.
Download colinear block data
The user can download a file containing colinear block information between two plants by choosing the two plants in the combo box and clicking the ‘download’ button. To decrease file size, the file is compressed by gzip, a popular file format that can easily be decompressed by many widely used programs. The file is written in comma-separated values (CSV) format and can be read and handled by most spreadsheet programs such as Microsoft Excel and Calc in LibreOffice (http://www.libreoffice.org/). The file contains not only gene pairs in colinear blocks but also additional data such as Ka and Ks values of the pairs.
CONCLUSIONS
To facilitate investigations into the functional and evolutionary consequences of gene and GD, we have determined and provided colinear blocks in plants from a single resource based on uniform standards. Many programs have been developed to determine colinear blocks, with different sensitivities and specificities in colinear block prediction (26,27). Among them, the current version of PGDD used MCScan, which shows a consistent, high accuracy prediction (27). PGDD has provided data used in much research (28–43,52–54) for past 5 years, and for the past 1 year alone, PGDD has been used by researchers from 111 countries with a total of 713 254 accession logs.
While continually adding new genome data to PGDD, we are also preparing new functions to enhance PGDD. For example, at present, users can access data in PGDD just by connecting to the web site or by downloading colinear block data files. To make it possible that other web services or programs can access PGDD data via the internet, we plan to add OpenAPI functions that enable web sites to interact with each other and build RESTful web services that make the data easily accessed over HTTP by clients. Besides developing the OpenAPI and RESTful web services, we plan to develop interfaces to link multiple data sources such as the VISTA (55) suite of programs and databases for multi-way analysis of genomic sequences, and CoGe (22), web application to display the homologous regions across multiple genomes. Hence, new functions and integration of multiple data sources are intended to further enhance the PGDD database as a platform to study many evolutionary questions.
FUNDING
A.H.P. appreciates funding from the National Science Foundation [NSF: DBI 0849896, MCB 0821096, MCB 1021718]; Resources and technical expertise from the University of Georgia (in part); Georgia Advanced Computing Resource Center, a partnership between the Office of the Vice President for Research and the Office of the Chief Information Officer. Funding for open access charge: NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 2.Hughes T, Roberts C, Dai H, Jones A, Meyer M, Slade D, Burchard J, Dow S, Ward T, Kidd M, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 4.Stephens S. Possible significance of duplications in evolution. Adv. Genet. 1951;4:247–265. doi: 10.1016/s0065-2660(08)60237-0. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium MGS. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Smith SF, Snell P, Gruetzner F, Bench AJ, Haaf T, Metcalfe JA, Green AR, Elgar G. Analyses of the extent of shared synteny and conserved gene orders between the genome of Fugu rubripes and human 20q. Genome Res. 2002;12:776–784. doi: 10.1101/gr.221802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proc. Natl Acad. Sci. USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang YW, Lai KN, Tai PY, Li WH. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 1999;48:597–604. doi: 10.1007/pl00006502. [DOI] [PubMed] [Google Scholar]
- 10.Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Genet. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowers JE, Chapman BA, Rong JK, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 13.Paterson AH, Freeling M, Sasaki T. Grains of knowledge: genomics of model cereals. Genome Res. 2005;15:1643–1650. doi: 10.1101/gr.3725905. [DOI] [PubMed] [Google Scholar]
- 14.Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annu. Rev. Plant Biol. 2010;61:349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- 15.Gu ZL, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 16.Christoffels A, Koh EGL, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- 17.Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- 18.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 19.Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M. The origins of eukaryotic gene structure. Mol. Biol. Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 21.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 22.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 23.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouard M, Guignon V, Aluome C, Laporte MA, Droc G, Walde C, Zmasek CM, Perin C, Conte MG. GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res. 2011;39:D1095–D1102. doi: 10.1093/nar/gkq811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012;158:590–600. doi: 10.1104/pp.111.189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proost S, Fostier J, De Witte D, Dhoedt B, Demeester P, Van de Peer Y, Vandepoele K. i-ADHoRe 3.0–fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012;40:e11. doi: 10.1093/nar/gkr955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Liu B, Yu L, Feng D, Wang H, Wang J. Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol. Biol. 2009;9:90. doi: 10.1186/1471-2148-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyun TK, Kim JS, Kwon SY, Kim SH. Comparative genomic analysis of mitogen activated protein kinase gene family in grapevine. Genes Genom. 2010;32:275–281. [Google Scholar]
- 30.Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One. 2010;5:e10065. doi: 10.1371/journal.pone.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Causier B, Castillo R, Xue Y, Schwarz-Sommer Z, Davies B. Tracing the evolution of the floral homeotic B- and C-function genes through genome synteny. Mol. Biol. Evol. 2010;27:2651–2664. doi: 10.1093/molbev/msq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmieri F, Pierri CL, De Grassi A, Nunes-Nesi A, Fernie AR. Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 2011;66:161–181. doi: 10.1111/j.1365-313X.2011.04516.x. [DOI] [PubMed] [Google Scholar]
- 33.Hwang SG, Kim DS, Jang CS. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica. 2011;139:1023–1032. doi: 10.1007/s10709-011-9604-y. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Zhang YM. Molecular evolution of glycinin and beta-conglycinin gene families in soybean (Glycine max L. Merr.) Heredity. 2011;106:633–641. doi: 10.1038/hdy.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe M, Mochida K, Kato T, Tabata S, Yoshimoto N, Noji M, Saito K. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. Plant Cell. 2008;20:2484–2496. doi: 10.1105/tpc.108.060335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopriva S, Mugford SG, Matthewman C, Koprivova A. Plant sulfate assimilation genes: redundancy versus specialization. Plant Cell Rep. 2009;28:1769–1780. doi: 10.1007/s00299-009-0793-0. [DOI] [PubMed] [Google Scholar]
- 37.Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl Acad. Sci. USA. 2009;106:7251–7256. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okazaki Y, Shimojima M, Sawada Y, Toyooka K, Narisawa T, Mochida K, Tanaka H, Matsuda F, Hirai A, Hirai MY, et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell. 2009;21:892–909. doi: 10.1105/tpc.108.063925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H, Bowers JE, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl Acad. Sci. USA. 2010;107:472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Wang X, Tang H, Tan X, Ficklin SP, Feltus FA, Paterson AH. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS One. 2011;6:e28150. doi: 10.1371/journal.pone.0028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makino T, McLysaght A. Positionally-biased gene loss after whole genome duplication: evidence from human, yeast and plant. Genome Res. 2012;22:2427–2435. doi: 10.1101/gr.131953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Wang X, Paterson AH. Genome and gene duplications and gene expression divergence: a view from plants. Ann. NY Acad. Sci. 2012;1256:1–14. doi: 10.1111/j.1749-6632.2011.06384.x. [DOI] [PubMed] [Google Scholar]
- 44.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzales MD, Archuleta E, Farmer A, Gajendran K, Grant D, Shoemaker R, Beavis WD, Waugh ME. The Legume Information System (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res. 2005;33:D660–D665. doi: 10.1093/nar/gki128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sjodin A, Street NR, Sandberg G, Gustafsson P, Jansson S. The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol. 2009;182:1013–1025. doi: 10.1111/j.1469-8137.2009.02807.x. [DOI] [PubMed] [Google Scholar]
- 47.D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- 48.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 51.Tang H, Bowers JE, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl Acad. Sci. USA. 2009;107:472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cannon SB, Shoemaker RC. Evolutionary and comparative analyses of the soybean genome. Breeding Sci. 2012;61:437–444. doi: 10.1270/jsbbs.61.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mochida K, Shinozaki K. Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol. 2010;51:497–523. doi: 10.1093/pcp/pcq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim C, Zhang D, Auckland SA, Rainville LK, Jakob K, Kronmiller B, Sacks EJ, Deuter M, Paterson AH. SSR-based genetic maps of Miscanthus sinensis and M. sacchariflorus, and their comparison to sorghum. Theor. Appl. Genet. 2012;124:1325–1338. doi: 10.1007/s00122-012-1790-1. [DOI] [PubMed] [Google Scholar]
- 55.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]